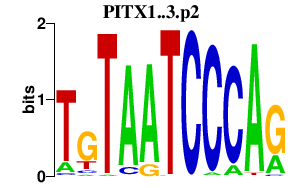
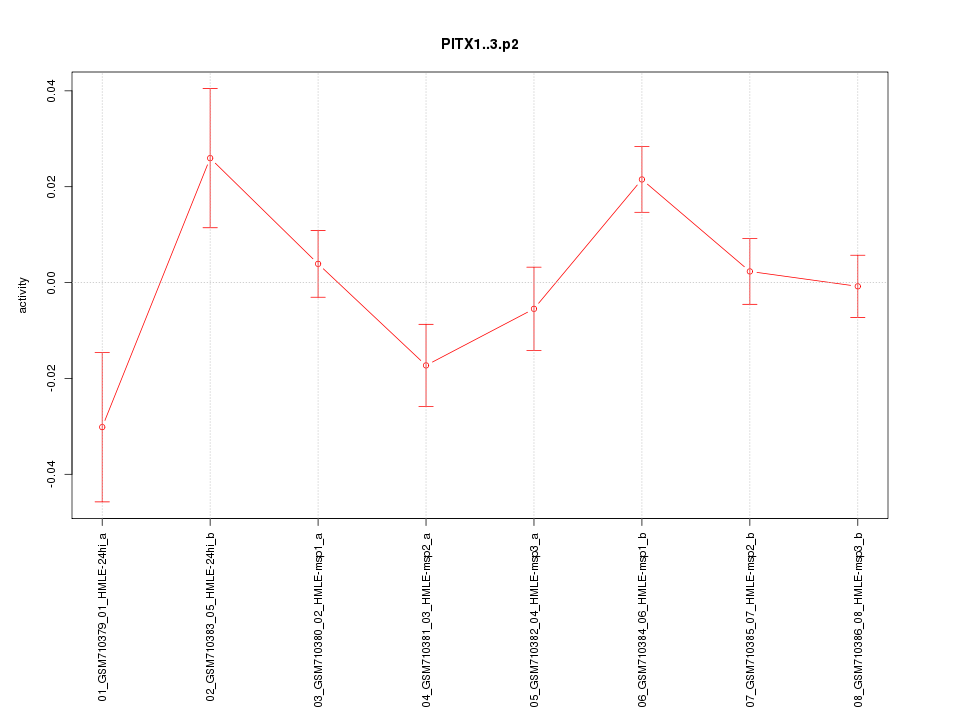
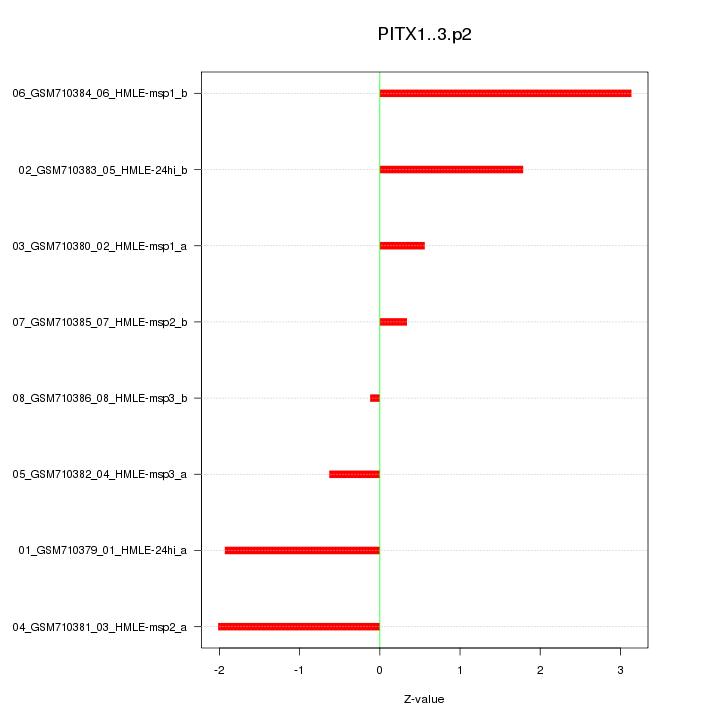
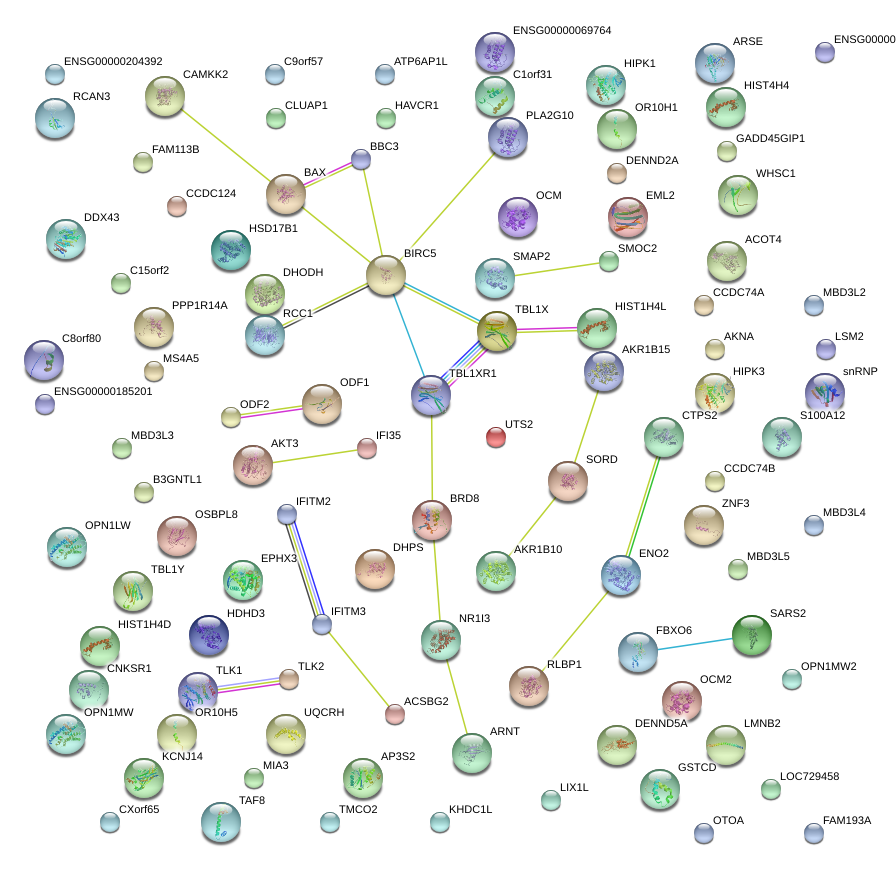
|
chr17_+_41158741
|
0.952
|
NM_005533
|
IFI35
|
interferon-induced protein 35
|
|
chr1_+_28832454
|
0.867
|
NM_001048199
|
RCC1
|
regulator of chromosome condensation 1
|
|
chrX_+_153448084
|
0.766
|
NM_000513
NM_001048181
|
OPN1MW
OPN1MW2
|
opsin 1 (cone pigments), medium-wave-sensitive
opsin 1 (cone pigments), medium-wave-sensitive 2
|
|
chrX_+_153485202
|
0.740
|
NM_000513
NM_001048181
|
OPN1MW
OPN1MW2
|
opsin 1 (cone pigments), medium-wave-sensitive
opsin 1 (cone pigments), medium-wave-sensitive 2
|
|
chr11_+_33363104
|
0.699
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr3_-_126327397
|
0.688
|
NM_001039783
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor
|
|
chr1_-_153348057
|
0.646
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr16_+_72042486
|
0.636
|
NM_001361
|
DHODH
|
dihydroorotate dehydrogenase (quinone)
|
|
chr12_+_7023490
|
0.626
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr12_-_124068864
|
0.620
|
|
|
|
|
chr1_-_244006885
|
0.611
|
NM_001206729
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr8_-_27941387
|
0.579
|
NM_001010906
|
C8orf80
|
chromosome 8 open reading frame 80
|
|
chr6_+_42018250
|
0.560
|
NM_138572
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr17_-_81006406
|
0.554
|
|
B3GNTL1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1
|
|
chrX_-_2882288
|
0.538
|
NM_000047
|
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1)
|
|
chr2_+_132285469
|
0.538
|
NM_138770
|
CCDC74A
|
coiled-coil domain containing 74A
|
|
chr1_+_247419273
|
0.537
|
NM_173858
|
VN1R5
|
vomeronasal 1 receptor 5 (gene/pseudogene)
|
|
chr1_-_16134192
|
0.522
|
NM_001089591
|
UQCRHL
|
ubiquinol-cytochrome c reductase hinge protein-like
|
|
chr19_+_7049349
|
0.518
|
NM_144614
|
MBD3L2
|
methyl-CpG binding domain protein 3-like 2
|
|
chr5_-_156486128
|
0.505
|
|
HAVCR1
|
hepatitis A virus cellular receptor 1
|
|
chr15_-_89764809
|
0.496
|
NM_000326
|
RLBP1
|
retinaldehyde binding protein 1
|
|
chr16_-_14788429
|
0.493
|
NM_003561
|
PLA2G10
|
phospholipase A2, group X
|
|
chr14_+_74058409
|
0.493
|
NM_152331
|
ACOT4
|
acyl-CoA thioesterase 4
|
|
chr12_-_121736110
|
0.489
|
NM_006549
NM_153499
NM_153500
NM_172214
NM_172215
NM_172216
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr19_-_13068033
|
0.488
|
NM_052850
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1
|
|
chr1_+_24840800
|
0.487
|
NM_001251980
NM_001251982
NM_001251985
|
RCAN3
|
RCAN family member 3
|
|
chr12_+_47610051
|
0.487
|
NM_138371
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr9_-_116138340
|
0.483
|
NM_031219
|
HDHD3
|
haloacid dehalogenase-like hydrolase domain containing 3
|
|
chr7_+_134212343
|
0.471
|
NM_020299
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase)
|
|
chr16_+_21695749
|
0.464
|
NM_001161683
|
OTOA
|
otoancorin
|
|
chr19_+_6135650
|
0.459
|
NM_030924
|
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2
|
|
chr19_-_23598867
|
0.454
|
|
|
|
|
chr6_+_42018264
|
0.453
|
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr16_+_21689834
|
0.453
|
NM_144672
|
OTOA
|
otoancorin
|
|
chr1_+_114496498
|
0.450
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr19_-_23598839
|
0.450
|
|
|
|
|
chr7_-_99680358
|
0.448
|
|
ZNF3
|
zinc finger protein 3
|
|
chr19_+_15904760
|
0.442
|
NM_001004466
|
OR10H5
|
olfactory receptor, family 10, subfamily H, member 5
|
|
chr6_+_74104575
|
0.440
|
|
DDX43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43
|
|
chr16_+_3559893
|
0.433
|
NM_024793
|
CLUAP1
|
clusterin associated protein 1
|
|
chr4_+_1894508
|
0.428
|
NM_007331
NM_133330
NM_133331
NM_133335
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr1_-_7973286
|
0.424
|
|
UTS2
|
urotensin 2
|
|
chr9_-_117156684
|
0.417
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr15_+_45315301
|
0.417
|
NM_003104
|
SORD
|
sorbitol dehydrogenase
|
|
chr1_+_40859016
|
0.416
|
NM_001198979
|
SMAP2
|
small ArfGAP2
|
|
chrX_+_9431314
|
0.415
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr19_-_38747171
|
0.413
|
NM_001243947
NM_033256
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A
|
|
chr19_+_48964505
|
0.413
|
NM_170720
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14
|
|
chr7_-_140302311
|
0.411
|
NM_015689
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr19_-_15343194
|
0.411
|
NM_024794
|
EPHX3
|
epoxide hydrolase 3
|
|
chr15_+_24920540
|
0.406
|
NM_018958
|
C15orf2
|
chromosome 15 open reading frame 2
|
|
chr5_+_81601165
|
0.405
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr1_+_40713572
|
0.405
|
NM_001008740
|
TMCO2
|
transmembrane and coiled-coil domains 2
|
|
chr6_-_26189303
|
0.405
|
NM_003539
|
HIST1H4D
|
histone cluster 1, H4d
|
|
chr1_-_161207879
|
0.405
|
NM_001077469
NM_001077470
NM_001077471
NM_001077472
NM_001077473
NM_001077474
NM_001077475
NM_001077476
NM_001077477
NM_001077478
NM_001077479
NM_001077480
NM_001077481
NM_001077482
NM_005122
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3
|
|
chr11_-_9225780
|
0.402
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr19_+_18045904
|
0.399
|
NM_138442
|
CCDC124
|
coiled-coil domain containing 124
|
|
chrX_-_16730669
|
0.396
|
NM_175859
NM_019857
|
CTPS2
|
CTP synthase II
|
|
chr1_+_26504024
|
0.390
|
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr17_+_76210329
|
0.388
|
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr4_+_2627158
|
0.387
|
NM_003704
|
FAM193A
|
family with sequence similarity 193, member A
|
|
chr6_-_73935174
|
0.386
|
NM_001126063
|
KHDC1L
|
KH homology domain containing 1-like
|
|
chr19_-_2456954
|
0.384
|
NM_032737
|
LMNB2
|
lamin B2
|
|
chr6_+_109450506
|
0.378
|
NM_173830
|
CEP57L1
|
centrosomal protein 57kDa-like 1
|
|
chr1_-_150849147
|
0.378
|
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr6_-_31774725
|
0.367
|
NM_021177
|
LSM2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr9_-_74675498
|
0.362
|
NM_001128618
|
C9orf57
|
chromosome 9 open reading frame 57
|
|
chr11_+_308106
|
0.362
|
NM_006435
|
IFITM2
|
interferon induced transmembrane protein 2 (1-8D)
|
|
chr2_-_171923482
|
0.361
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr19_+_49458031
|
0.358
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr19_-_39443326
|
0.356
|
NM_148169
|
FBXO17
|
F-box protein 17
|
|
chr19_+_49458129
|
0.355
|
|
BAX
|
BCL2-associated X protein
|
|
chr7_-_97619350
|
0.346
|
NM_006188
|
OCM2
|
oncomodulin 2
|
|
chr4_+_106631966
|
0.344
|
NM_001031720
|
GSTCD
|
glutathione S-transferase, C-terminal domain containing
|
|
chr11_+_60197061
|
0.338
|
NM_023945
|
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5
|
|
chr19_-_47735858
|
0.333
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr1_+_11724148
|
0.329
|
NM_018438
|
FBXO6
|
F-box protein 6
|
|
chr19_-_46142643
|
0.328
|
|
EML2
|
echinoderm microtubule associated protein like 2
|
|
chr15_-_23115243
|
0.325
|
|
LOC283683
|
uncharacterized LOC283683
|
|
chrX_-_70326637
|
0.321
|
NM_001025265
|
CXorf65
|
chromosome X open reading frame 65
|
|
chr1_+_145487301
|
0.318
|
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr5_+_157184338
|
0.318
|
|
|
|
|
chr17_+_40703983
|
0.316
|
NM_000413
|
HSD17B1
|
hydroxysteroid (17-beta) dehydrogenase 1
|
|
chr1_+_234509213
|
0.316
|
NM_001206641
NM_001012985
|
C1orf31
|
chromosome 1 open reading frame 31
|
|
chr19_-_12792676
|
0.315
|
|
DHPS
|
deoxyhypusine synthase
|
|
chr15_-_90437616
|
0.314
|
NM_005829
|
AP3S2
|
adaptor-related protein complex 3, sigma 2 subunit
|
|
chr9_+_131217433
|
0.312
|
NM_153436
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr19_-_12792577
|
0.312
|
|
DHPS
|
deoxyhypusine synthase
|
|
chr17_+_17380264
|
0.310
|
NM_018019
|
MED9
|
mediator complex subunit 9
|
|
chr8_+_11627144
|
0.308
|
NM_001135746
NM_001135747
NM_001135748
NM_145043
|
NEIL2
|
nei endonuclease VIII-like 2 (E. coli)
|
|
chr1_-_202896315
|
0.307
|
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr18_+_61370193
|
0.304
|
NM_080475
|
SERPINB11
|
serpin peptidase inhibitor, clade B (ovalbumin), member 11 (gene/pseudogene)
|
|
chr8_-_38008332
|
0.304
|
NM_000349
|
STAR
|
steroidogenic acute regulatory protein
|
|
chr10_+_105005643
|
0.301
|
NM_001143909
|
LOC729020
|
rcRPE
|
|
chr22_-_42343079
|
0.300
|
NM_001002876
NM_024053
|
CENPM
|
centromere protein M
|
|
chr12_-_21757752
|
0.298
|
NM_021957
|
GYS2
|
glycogen synthase 2 (liver)
|
|
chr21_-_31859666
|
0.294
|
NM_181608
|
KRTAP19-2
|
keratin associated protein 19-2
|
|
chr22_-_17646176
|
0.294
|
NM_017829
|
CECR5
|
cat eye syndrome chromosome region, candidate 5
|
|
chr21_-_39493349
|
0.293
|
NM_005867
|
DSCR4
|
Down syndrome critical region gene 4
|
|
chr17_+_71229357
|
0.293
|
|
C17orf80
|
chromosome 17 open reading frame 80
|
|
chr16_+_12058963
|
0.292
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr2_+_75059781
|
0.291
|
NM_000189
|
HK2
|
hexokinase 2
|
|
chrX_-_129272014
|
0.291
|
NM_001130846
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr17_-_6947241
|
0.290
|
NM_153357
|
SLC16A11
|
solute carrier family 16, member 11 (monocarboxylic acid transporter 11)
|
|
chr18_+_3453771
|
0.290
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr1_-_27701302
|
0.289
|
NM_003665
NM_173452
|
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 (Hakata antigen)
|
|
chrX_+_96138906
|
0.288
|
NM_013347
|
RPA4
|
replication protein A4, 30kDa
|
|
chr5_-_156485118
|
0.288
|
|
HAVCR1
|
hepatitis A virus cellular receptor 1
|
|
chr19_-_42931500
|
0.287
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr19_-_46142662
|
0.287
|
NM_012155
|
EML2
|
echinoderm microtubule associated protein like 2
|
|
chr19_+_17530841
|
0.282
|
|
FAM125A
|
family with sequence similarity 125, member A
|
|
chr11_+_68367824
|
0.280
|
|
PPP6R3
|
protein phosphatase 6, regulatory subunit 3
|
|
chr1_-_150849211
|
0.279
|
NM_001197325
NM_001668
NM_178427
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr19_-_12792698
|
0.278
|
NM_001930
NM_013406
|
DHPS
|
deoxyhypusine synthase
|
|
chr22_+_21128378
|
0.277
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr20_+_51801821
|
0.276
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr22_-_42336222
|
0.273
|
NM_001110215
|
CENPM
|
centromere protein M
|
|
chr11_-_67211291
|
0.273
|
NM_001018070
|
CORO1B
|
coronin, actin binding protein, 1B
|
|
chr9_+_132388451
|
0.273
|
|
METTL11A
|
methyltransferase like 11A
|
|
chr5_+_54320083
|
0.272
|
NM_002104
|
GZMK
|
granzyme K (granzyme 3; tryptase II)
|
|
chr20_+_33462765
|
0.272
|
NM_001242393
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr1_+_3668961
|
0.271
|
NM_152492
|
CCDC27
|
coiled-coil domain containing 27
|
|
chrX_+_48620153
|
0.270
|
NM_001080489
|
GLOD5
|
glyoxalase domain containing 5
|
|
chr9_+_132388404
|
0.269
|
NM_014064
|
METTL11A
|
methyltransferase like 11A
|
|
chr11_-_62607627
|
0.267
|
NM_018093
|
WDR74
|
WD repeat domain 74
|
|
chr20_-_39928708
|
0.267
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr19_+_10812111
|
0.266
|
NM_031209
|
QTRT1
|
queuine tRNA-ribosyltransferase 1
|
|
chr12_-_49177876
|
0.265
|
NM_015270
|
ADCY6
|
adenylate cyclase 6
|
|
chr3_+_109128836
|
0.264
|
NM_001145553
|
FLJ25363
|
uncharacterized LOC401082
|
|
chr2_-_130902568
|
0.263
|
NM_207310
|
CCDC74B
|
coiled-coil domain containing 74B
|
|
chr19_+_14551080
|
0.262
|
NM_213560
|
PKN1
|
protein kinase N1
|
|
chr16_+_32752634
|
0.261
|
|
LOC100289574
|
hect domain and RLD 2 pseudogene
|
|
chr7_+_76054251
|
0.260
|
NM_001110354
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor)
|
|
chr6_-_31763551
|
0.260
|
NM_006295
|
VARS
|
valyl-tRNA synthetase
|
|
chr15_-_70878943
|
0.259
|
|
|
|
|
chr2_+_204571197
|
0.259
|
NM_001243077
NM_001243078
NM_006139
|
CD28
|
CD28 molecule
|
|
chr10_-_99393851
|
0.258
|
NM_001098831
|
MORN4
|
MORN repeat containing 4
|
|
chr9_-_117160748
|
0.256
|
|
AKNA
|
AT-hook transcription factor
|
|
chr1_+_28832552
|
0.256
|
|
SNHG3
|
small nucleolar RNA host gene 3 (non-protein coding)
|
|
chr19_-_17932312
|
0.256
|
NM_005543
|
INSL3
|
insulin-like 3 (Leydig cell)
|
|
chr10_-_105452878
|
0.256
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr19_-_51893791
|
0.256
|
NM_001193623
|
LOC147646
|
uncharacterized LOC147646
|
|
chr16_-_25026650
|
0.256
|
NM_001006634
NM_018054
|
ARHGAP17
|
Rho GTPase activating protein 17
|
|
chr17_-_8113862
|
0.250
|
NM_004217
|
AURKB
|
aurora kinase B
|
|
chr16_-_32199406
|
0.248
|
|
LOC100289574
|
hect domain and RLD 2 pseudogene
|
|
chr5_-_156485969
|
0.248
|
NM_001173393
|
HAVCR1
|
hepatitis A virus cellular receptor 1
|
|
chr1_+_175036993
|
0.248
|
NM_022093
|
TNN
|
tenascin N
|
|
chr13_-_45992454
|
0.247
|
|
SLC25A30
|
solute carrier family 25, member 30
|
|
chr1_-_109203663
|
0.247
|
NM_001102592
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis)
|
|
chr11_+_118955569
|
0.247
|
NM_000190
|
HMBS
|
hydroxymethylbilane synthase
|
|
chr12_-_31945174
|
0.246
|
NM_001013699
|
H3F3C
|
H3 histone, family 3C
|
|
chr4_-_159956332
|
0.246
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr1_-_182359413
|
0.246
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr5_+_37379382
|
0.245
|
|
WDR70
|
WD repeat domain 70
|
|
chr19_+_14551052
|
0.243
|
|
PKN1
|
protein kinase N1
|
|
chr11_+_65101224
|
0.243
|
NM_006268
|
DPF2
|
D4, zinc and double PHD fingers family 2
|
|
chrX_+_2959511
|
0.243
|
NM_001201539
|
ARSF
|
arylsulfatase F
|
|
chr16_-_15833937
|
0.243
|
|
MYH11
|
myosin, heavy chain 11, smooth muscle
|
|
chr12_-_46357949
|
0.242
|
|
|
|
|
chr14_-_70826303
|
0.241
|
|
COX16
|
COX16 cytochrome c oxidase assembly homolog (S. cerevisiae)
|
|
chr14_-_35008781
|
0.240
|
NM_018453
|
EAPP
|
E2F-associated phosphoprotein
|
|
chr6_+_35995583
|
0.238
|
|
MAPK14
|
mitogen-activated protein kinase 14
|
|
chr22_+_39795745
|
0.237
|
NM_006116
NM_153497
|
TAB1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1
|
|
chr19_+_6361558
|
0.236
|
|
CLPP
|
ClpP caseinolytic peptidase, ATP-dependent, proteolytic subunit homolog (E. coli)
|
|
chr13_-_21348081
|
0.235
|
|
N6AMT2
|
N-6 adenine-specific DNA methyltransferase 2 (putative)
|
|
chr1_+_151253977
|
0.235
|
|
ZNF687
|
zinc finger protein 687
|
|
chr6_+_31598366
|
0.234
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr16_-_1922079
|
0.234
|
NM_152764
|
C16orf73
|
chromosome 16 open reading frame 73
|
|
chr10_+_35535952
|
0.233
|
NM_181698
|
CCNY
|
cyclin Y
|
|
chr19_+_14551087
|
0.232
|
|
PKN1
|
protein kinase N1
|
|
chr19_+_7745706
|
0.230
|
NM_001042462
|
TRAPPC5
|
trafficking protein particle complex 5
|
|
chr9_-_116139148
|
0.230
|
|
HDHD3
|
haloacid dehalogenase-like hydrolase domain containing 3
|
|
chr17_-_56358161
|
0.230
|
|
MPO
|
myeloperoxidase
|
|
chr11_+_5641173
|
0.229
|
NM_001003827
|
TRIM34
|
tripartite motif containing 34
|
|
chr17_-_73761216
|
0.228
|
NM_000154
|
GALK1
|
galactokinase 1
|
|
chr11_-_68518944
|
0.225
|
NM_001039656
NM_004923
|
MTL5
|
metallothionein-like 5, testis-specific (tesmin)
|
|
chr16_-_1922161
|
0.224
|
NM_001163560
|
C16orf73
|
chromosome 16 open reading frame 73
|
|
chr6_-_109762373
|
0.223
|
NM_001111298
NM_173672
|
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6
|
|
chr10_+_126490353
|
0.223
|
NM_032182
|
FAM175B
|
family with sequence similarity 175, member B
|
|
chr19_-_13068008
|
0.222
|
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1
|
|
chr5_+_132406080
|
0.222
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr1_-_150849062
|
0.221
|
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr16_+_19078958
|
0.220
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr13_-_21348054
|
0.220
|
NM_174928
|
N6AMT2
|
N-6 adenine-specific DNA methyltransferase 2 (putative)
|
|
chr2_+_85843247
|
0.219
|
|
USP39
|
ubiquitin specific peptidase 39
|
|
chr13_+_41363546
|
0.218
|
NM_014252
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15
|
|
chr12_-_122296619
|
0.218
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr13_+_41363642
|
0.218
|
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15
|
|
chr1_+_24834083
|
0.217
|
NM_001251978
|
RCAN3
|
RCAN family member 3
|
|
chr1_-_713985
|
0.216
|
|
LOC100288069
|
general transcription factor IIi pseudogene
|
|
chr20_+_361272
|
0.216
|
NM_021158
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr7_+_76026819
|
0.215
|
NM_007155
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor)
|
|
chr2_+_97001463
|
0.215
|
NM_015341
|
NCAPH
|
non-SMC condensin I complex, subunit H
|
|
chr17_-_5372235
|
0.214
|
NM_001199699
NM_020162
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33
|
|
chrX_+_35816458
|
0.214
|
NM_001099921
|
MAGEB16
|
melanoma antigen family B, 16
|
|
chr12_-_123706393
|
0.214
|
NM_022782
|
MPHOSPH9
|
M-phase phosphoprotein 9
|
|
chr17_-_56358266
|
0.213
|
NM_000250
|
MPO
|
myeloperoxidase
|
|
chr4_+_70861647
|
0.213
|
NM_001009181
NM_003154
|
STATH
|
statherin
|
|
chr6_-_31107110
|
0.212
|
NM_014069
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2
|
|
chr6_+_31430955
|
0.211
|
|
HCP5
|
HLA complex P5 (non-protein coding)
|