|
chr6_-_11779279
|
2.139
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chrX_-_24665454
|
1.499
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr5_+_36608430
|
1.445
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr18_-_53089722
|
1.252
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chrX_-_24665225
|
1.152
|
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr10_-_98031154
|
1.132
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr6_+_26124372
|
1.099
|
NM_003512
|
HIST1H2AC
|
histone cluster 1, H2ac
|
|
chr10_-_123356158
|
1.081
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr17_-_56494889
|
1.059
|
|
RNF43
|
ring finger protein 43
|
|
chr9_-_140196702
|
1.042
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr1_+_206730456
|
1.015
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr12_-_31479120
|
1.014
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr12_-_31479084
|
0.994
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr12_+_107712151
|
0.976
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chrX_-_117119282
|
0.961
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr12_-_31479038
|
0.950
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr9_-_117160748
|
0.944
|
|
AKNA
|
AT-hook transcription factor
|
|
chr20_+_62795773
|
0.943
|
|
MYT1
|
myelin transcription factor 1
|
|
chr6_+_27215479
|
0.927
|
NM_005865
|
PRSS16
|
protease, serine, 16 (thymus)
|
|
chr17_-_56494930
|
0.911
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chr6_+_26183957
|
0.904
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr7_+_16793320
|
0.879
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr6_-_99797530
|
0.878
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr1_+_206730497
|
0.835
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr11_+_34642587
|
0.818
|
NM_001206615
NM_012153
|
EHF
|
ets homologous factor
|
|
chr8_+_56792345
|
0.752
|
NM_001111097
NM_002350
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr8_+_56792392
|
0.742
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr7_-_41742666
|
0.740
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr6_-_26124130
|
0.736
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr12_+_113344738
|
0.733
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr8_+_56792399
|
0.719
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr1_+_156119734
|
0.709
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr5_+_145317297
|
0.695
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr4_-_40517913
|
0.672
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr1_-_228645517
|
0.668
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr6_+_37137882
|
0.665
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr6_-_170599568
|
0.642
|
NM_005618
|
DLL1
|
delta-like 1 (Drosophila)
|
|
chr2_+_207804277
|
0.627
|
NM_173077
|
CPO
|
carboxypeptidase O
|
|
chr1_+_6052699
|
0.601
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr6_+_26251826
|
0.598
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr3_+_186648273
|
0.598
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr16_+_23847271
|
0.592
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr1_+_6052758
|
0.584
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr2_-_113594325
|
0.579
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chrX_+_69674914
|
0.542
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr6_+_43139201
|
0.537
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr12_+_113344811
|
0.524
|
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr6_-_27114577
|
0.518
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr6_-_27100528
|
0.513
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr6_-_143266283
|
0.511
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr8_+_86099909
|
0.511
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr13_+_97794050
|
0.500
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr11_+_64073040
|
0.483
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr8_+_123793988
|
0.473
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr21_-_32201926
|
0.468
|
NM_181606
|
KRTAP7-1
|
keratin associated protein 7-1 (gene/pseudogene)
|
|
chr12_-_57328044
|
0.463
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr2_-_165551178
|
0.458
|
|
COBLL1
|
COBL-like 1
|
|
chr6_-_10415438
|
0.457
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr17_-_7307413
|
0.443
|
NM_152766
|
C17orf61-PLSCR3
C17orf61
|
C17orf61-PLSCR3 readthrough
chromosome 17 open reading frame 61
|
|
chr12_-_50616425
|
0.407
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr16_+_23847321
|
0.397
|
|
PRKCB
|
protein kinase C, beta
|
|
chr1_-_94586678
|
0.395
|
NM_000350
|
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4
|
|
chr8_+_123793894
|
0.390
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr6_+_26158332
|
0.383
|
NM_021063
NM_138720
|
HIST1H2BD
|
histone cluster 1, H2bd
|
|
chr1_+_149822627
|
0.382
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr1_-_149814264
|
0.377
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr6_+_43139190
|
0.369
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr17_+_9745847
|
0.369
|
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr1_-_154832187
|
0.358
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr19_-_1132109
|
0.354
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr1_-_149399159
|
0.350
|
NM_001024599
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr11_+_34642651
|
0.349
|
|
EHF
|
ets homologous factor
|
|
chr22_+_22758538
|
0.345
|
|
|
|
|
chr4_+_5526882
|
0.344
|
NM_005750
|
C4orf6
|
chromosome 4 open reading frame 6
|
|
chr1_-_149783910
|
0.342
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
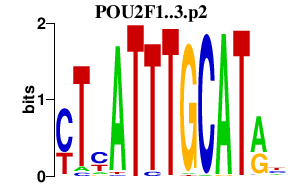
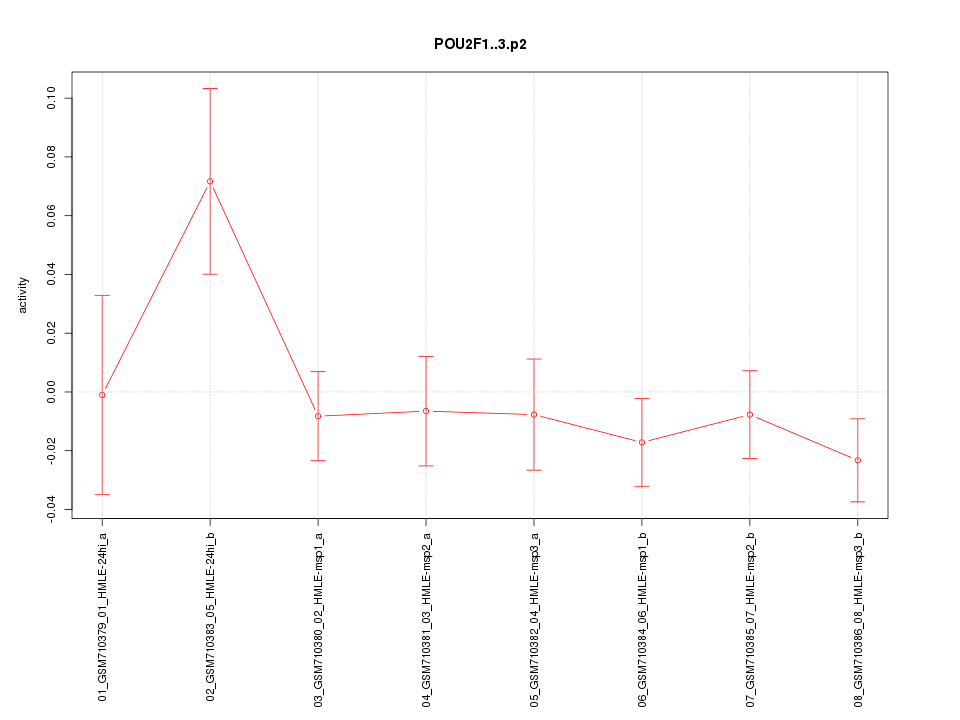
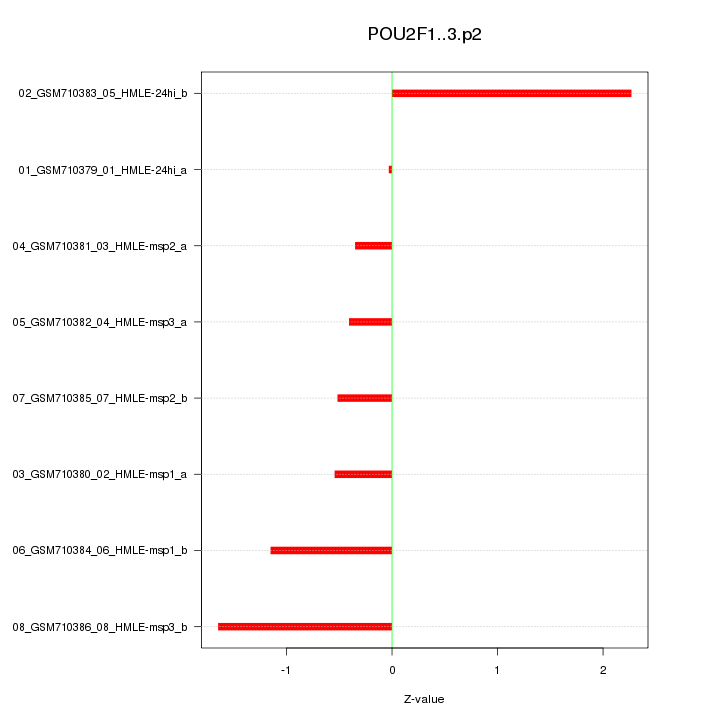
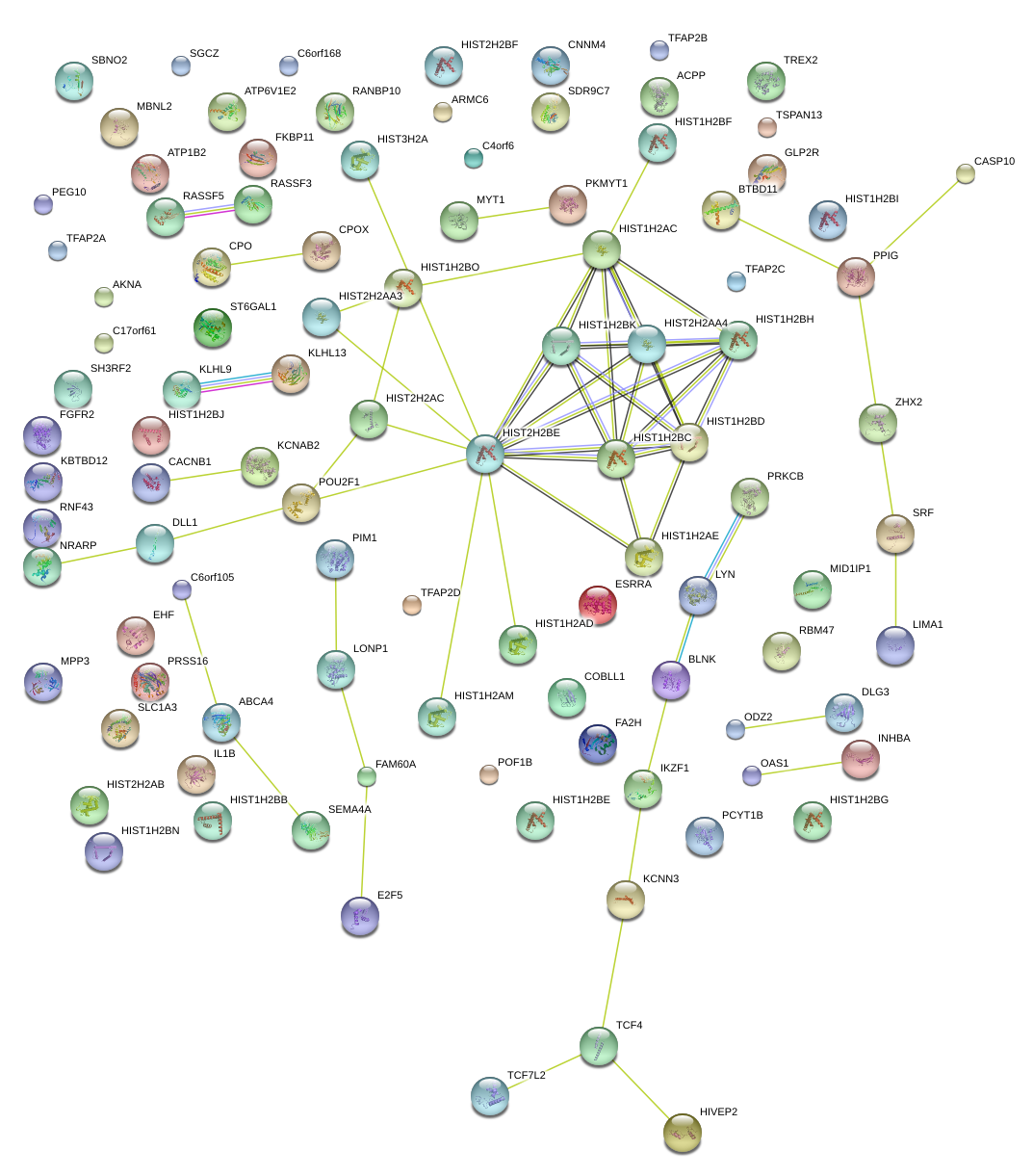
chr1_+_167298382
|
0.326
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr7_+_50348255
|
0.321
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chrX_-_84634742
|
0.318
|
NM_024921
|
POF1B
|
premature ovarian failure, 1B
|
|
chr2_+_202047620
|
0.314
|
NM_001206524
NM_001206542
NM_001230
NM_032974
NM_032976
NM_032977
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr1_-_149859465
|
0.314
|
NM_175065
|
HIST2H2AB
HIST2H2BE
|
histone cluster 2, H2ab
histone cluster 2, H2be
|
|
chr8_+_123794043
|
0.309
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr17_+_7554239
|
0.305
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr2_+_97426571
|
0.301
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr6_+_26273143
|
0.298
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr19_+_19144617
|
0.291
|
|
ARMC6
|
armadillo repeat containing 6
|
|
chr5_+_166711842
|
0.291
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr3_+_127641901
|
0.289
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chrX_+_38660653
|
0.287
|
NM_001098790
NM_001098791
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr7_+_94292737
|
0.287
|
|
PEG10
|
paternally expressed 10
|
|
chr2_-_46747095
|
0.284
|
NM_080653
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2
|
|
chr3_+_132036276
|
0.282
|
|
ACPP
|
acid phosphatase, prostate
|
|
chr19_+_19144520
|
0.279
|
|
ARMC6
|
armadillo repeat containing 6
|
|
chr16_-_74808699
|
0.277
|
NM_024306
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr8_-_15095791
|
0.275
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr2_-_152684933
|
0.274
|
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr7_+_18535351
|
0.268
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr2_-_237172987
|
0.268
|
NM_212556
|
ASB18
|
ankyrin repeat and SOCS box containing 18
|
|
chr1_+_81771844
|
0.264
|
|
LPHN2
|
latrophilin 2
|
|
chr22_+_24033047
|
0.264
|
NM_153615
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4
|
|
chr18_-_52988916
|
0.264
|
NM_001243234
NM_001243235
|
TCF4
|
transcription factor 4
|
|
chr20_+_9198036
|
0.263
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr22_+_21986968
|
0.262
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr11_+_27062508
|
0.262
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr2_+_202047803
|
0.257
|
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr12_-_11139510
|
0.255
|
NM_176890
|
TAS2R50
|
taste receptor, type 2, member 50
|
|
chr19_+_41305100
|
0.254
|
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr3_+_132036210
|
0.254
|
NM_001099
NM_001134194
|
ACPP
|
acid phosphatase, prostate
|
|
chr17_+_7554415
|
0.253
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr3_+_151591428
|
0.252
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr16_-_74808474
|
0.251
|
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr7_-_141673572
|
0.248
|
NM_176817
|
TAS2R38
|
taste receptor, type 2, member 38
|
|
chrX_+_38660692
|
0.248
|
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr6_-_26216871
|
0.246
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr3_-_165555167
|
0.246
|
|
BCHE
|
butyrylcholinesterase
|
|
chr2_+_16080559
|
0.244
|
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr11_+_112047088
|
0.244
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr1_-_149858114
|
0.244
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr6_-_138820580
|
0.242
|
|
|
|
|
chr19_+_41305041
|
0.242
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr11_-_62369213
|
0.241
|
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr2_-_215896809
|
0.241
|
NM_015657
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr12_-_6484389
|
0.238
|
NM_001159576
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chrX_-_131547536
|
0.236
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr19_+_19144370
|
0.235
|
NM_001199196
NM_033415
|
ARMC6
|
armadillo repeat containing 6
|
|
chr12_-_48963802
|
0.235
|
NM_002289
|
LALBA
|
lactalbumin, alpha-
|
|
chr10_+_123922940
|
0.228
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr15_-_64995346
|
0.227
|
|
OAZ2
|
ornithine decarboxylase antizyme 2
|
|
chr2_-_152684689
|
0.226
|
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr1_-_186649421
|
0.222
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr12_-_11062160
|
0.221
|
NM_023920
|
TAS2R13
|
taste receptor, type 2, member 13
|
|
chr6_+_27861202
|
0.220
|
NM_003527
|
HIST1H2BO
|
histone cluster 1, H2bo
|
|
chrX_-_84634664
|
0.220
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr6_-_43655514
|
0.219
|
|
MRPS18A
|
mitochondrial ribosomal protein S18A
|
|
chr14_+_65007418
|
0.219
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr19_-_50169021
|
0.218
|
NM_001197122
NM_001197123
NM_001197124
NM_001197125
NM_001197126
NM_001197127
NM_001197128
NM_001571
|
IRF3
|
interferon regulatory factor 3
|
|
chr12_+_122064448
|
0.217
|
NM_032790
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1
|
|
chr6_-_43655539
|
0.214
|
NM_001193343
NM_018135
|
MRPS18A
|
mitochondrial ribosomal protein S18A
|
|
chr11_-_62369097
|
0.213
|
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr19_+_50169080
|
0.210
|
|
BCL2L12
|
BCL2-like 12 (proline rich)
|
|
chr22_+_32750871
|
0.209
|
NM_006604
|
RFPL3
|
ret finger protein-like 3
|
|
chr10_-_62332666
|
0.209
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr19_+_17337012
|
0.207
|
NM_024578
|
OCEL1
|
occludin/ELL domain containing 1
|
|
chr11_-_64084980
|
0.206
|
NM_016404
|
TRMT112
|
tRNA methyltransferase 11-2 homolog (S. cerevisiae)
|
|
chr9_-_2844098
|
0.205
|
NM_014878
|
KIAA0020
|
KIAA0020
|
|
chr14_-_23445885
|
0.204
|
|
AJUBA
|
ajuba LIM protein
|
|
chr19_-_40791291
|
0.201
|
NM_001243027
NM_001243028
NM_001626
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr9_-_3223007
|
0.200
|
|
|
|
|
chr11_+_46383115
|
0.199
|
NM_001105540
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr3_+_50192420
|
0.198
|
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr3_-_165555252
|
0.197
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr11_-_62358969
|
0.196
|
NM_022830
|
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific
|
|
chr11_-_62369250
|
0.194
|
NM_004739
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr10_-_97321094
|
0.194
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr11_-_115375065
|
0.193
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr1_+_41157241
|
0.193
|
NM_001142587
NM_001142588
NM_001142589
NM_014223
|
NFYC
|
nuclear transcription factor Y, gamma
|
|
chr19_-_50168866
|
0.193
|
|
IRF3
|
interferon regulatory factor 3
|
|
chr6_-_138820578
|
0.192
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr14_+_65007163
|
0.191
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr18_+_77866914
|
0.190
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr17_-_48072566
|
0.187
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr1_+_228645807
|
0.186
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr11_-_3013550
|
0.185
|
|
NAP1L4
|
nucleosome assembly protein 1-like 4
|
|
chr9_-_2844083
|
0.184
|
|
KIAA0020
|
KIAA0020
|
|
chr10_+_123923104
|
0.180
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr11_+_537521
|
0.179
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr19_-_19281090
|
0.178
|
NM_001145785
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr16_-_30107492
|
0.178
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr10_+_106113513
|
0.178
|
NM_001008723
|
CCDC147
|
coiled-coil domain containing 147
|
|
chr7_-_100183753
|
0.177
|
NM_002319
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr4_-_47983430
|
0.177
|
NM_001142564
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr1_+_167298280
|
0.176
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr6_+_31588460
|
0.175
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr1_-_68962679
|
0.174
|
|
DEPDC1
|
DEP domain containing 1
|
|
chr15_-_40600065
|
0.174
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr19_-_19281062
|
0.174
|
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr10_-_72648540
|
0.173
|
NM_000281
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha
|
|
chrX_+_123234462
|
0.173
|
|
STAG2
|
stromal antigen 2
|
|
chr7_-_44122083
|
0.170
|
|
POLM
|
polymerase (DNA directed), mu
|
|
chr7_-_44122036
|
0.169
|
|
POLM
|
polymerase (DNA directed), mu
|
|
chr6_-_134373618
|
0.168
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr3_-_121379701
|
0.168
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr6_+_143999058
|
0.166
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr22_+_23054862
|
0.165
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr7_+_16566504
|
0.164
|
NM_001195280
|
LRRC72
|
leucine rich repeat containing 72
|
|
chr18_-_24442534
|
0.163
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr21_-_39493349
|
0.161
|
NM_005867
|
DSCR4
|
Down syndrome critical region gene 4
|
|
chr6_+_31588444
|
0.161
|
NM_004638
NM_080686
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr18_+_44526786
|
0.161
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr2_+_16080639
|
0.160
|
NM_005378
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr18_+_76740274
|
0.160
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr5_-_142065586
|
0.159
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr5_-_2751768
|
0.158
|
NM_001134222
NM_033267
|
IRX2
|
iroquois homeobox 2
|
|
chr6_+_148663953
|
0.157
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr5_+_52776408
|
0.156
|
|
FST
|
follistatin
|
|
chr8_+_26435590
|
0.155
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr5_+_52776263
|
0.155
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr19_+_41305333
|
0.152
|
NM_053046
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr6_+_31588524
|
0.152
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr6_+_148663728
|
0.152
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr13_-_70682458
|
0.151
|
NM_020866
|
KLHL1
|
kelch-like 1 (Drosophila)
|