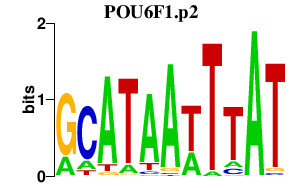
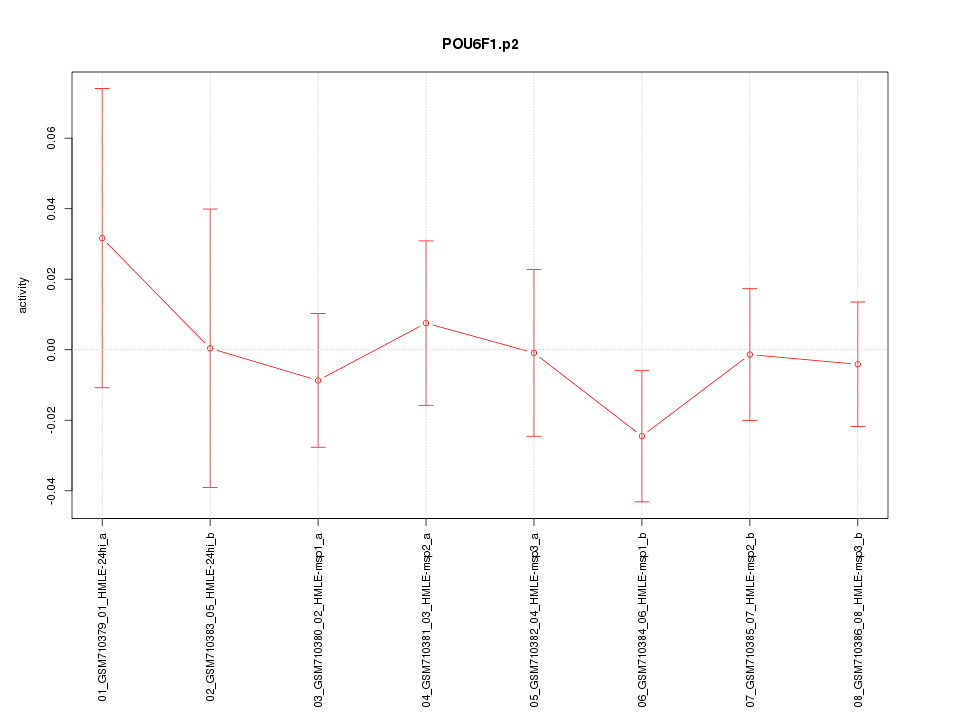
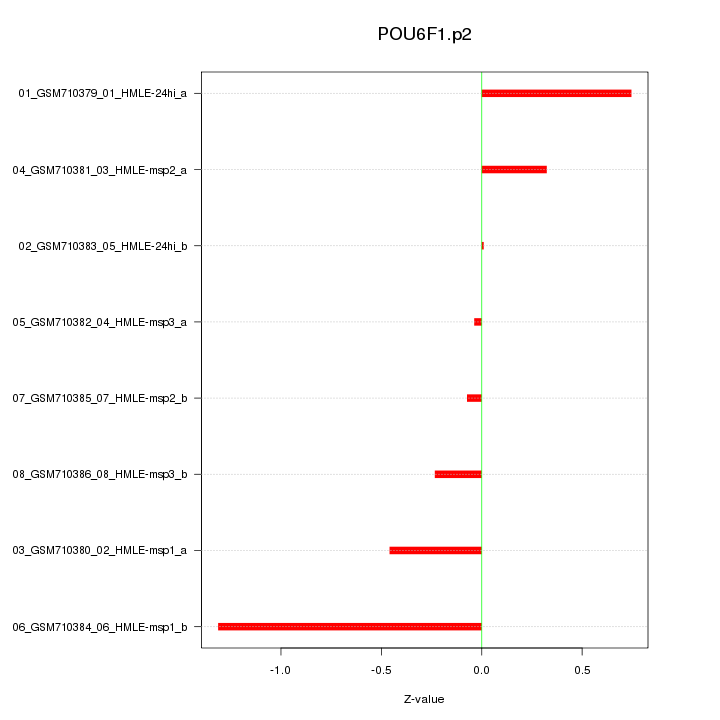
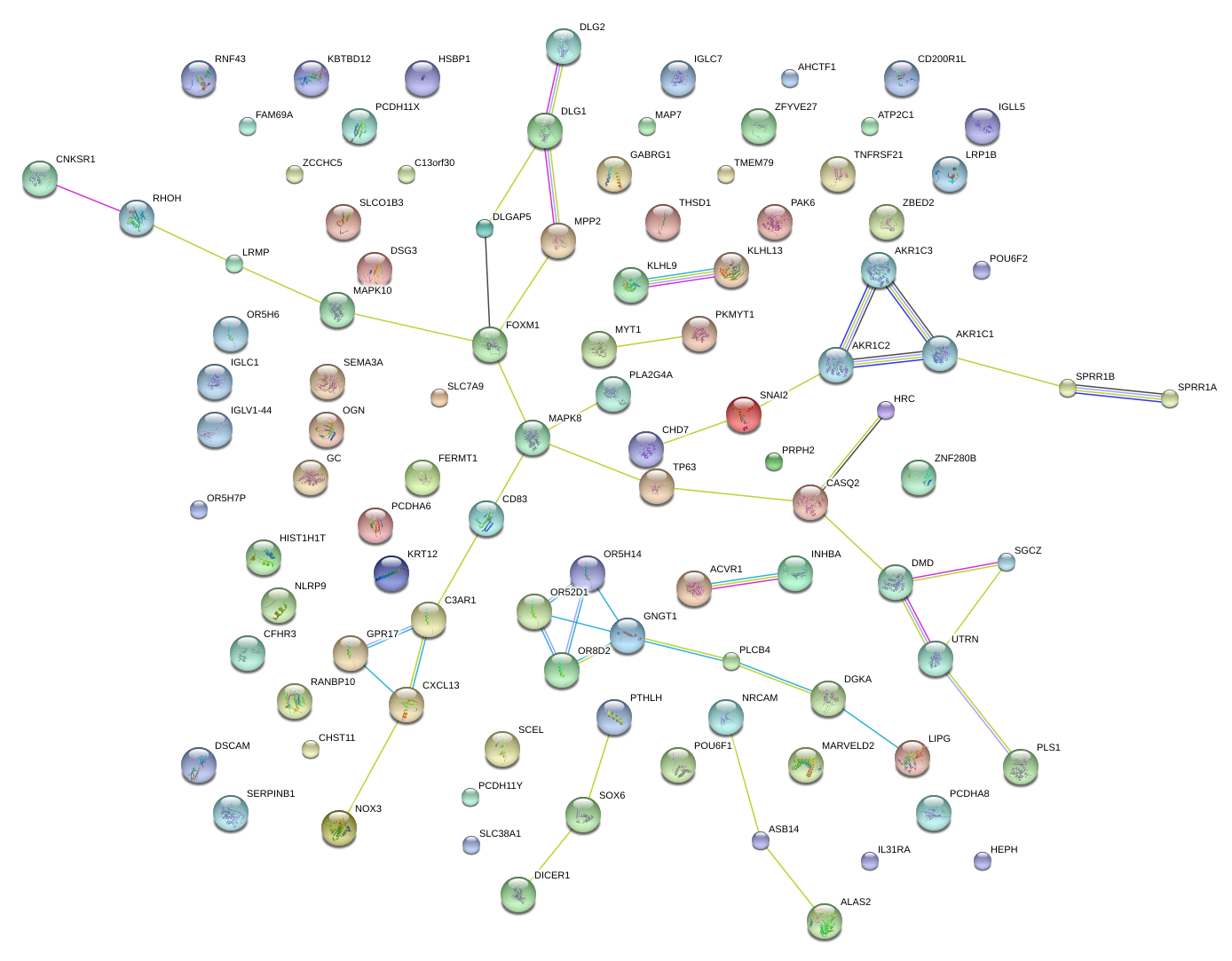
|
chr1_+_153003677
|
0.811
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr20_+_62795773
|
0.681
|
|
MYT1
|
myelin transcription factor 1
|
|
chr18_+_47088400
|
0.539
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr7_-_83824216
|
0.459
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chrX_+_65382432
|
0.399
|
NM_138737
|
HEPH
|
hephaestin
|
|
chr5_+_140220811
|
0.366
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr12_-_28124915
|
0.356
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr1_-_116311330
|
0.320
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr6_-_136847078
|
0.314
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr1_-_116311161
|
0.310
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr1_-_116311398
|
0.304
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr3_-_111314181
|
0.288
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr10_+_99498233
|
0.273
|
NM_001002261
|
ZFYVE27
|
zinc finger, FYVE domain containing 27
|
|
chr3_-_112564789
|
0.267
|
NM_001008784
NM_001199215
|
CD200R1L
|
CD200 receptor 1-like
|
|
chr19_-_33360641
|
0.264
|
NM_001126335
NM_001243036
NM_014270
|
SLC7A9
|
solute carrier family 7 (glycoprotein-associated amino acid transporter light chain, bo,+ system), member 9
|
|
chr9_-_95166827
|
0.263
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr17_-_56492938
|
0.237
|
|
RNF43
|
ring finger protein 43
|
|
chr1_+_15272414
|
0.234
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr3_+_127641901
|
0.225
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr1_+_186798031
|
0.225
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr4_+_40198526
|
0.216
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr10_+_5005685
|
0.204
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr7_-_107880492
|
0.203
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr11_-_16497917
|
0.203
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr6_-_138189313
|
0.192
|
|
|
|
|
chrX_-_117119282
|
0.186
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr12_+_105153258
|
0.181
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chrX_-_33229428
|
0.167
|
NM_004006
|
DMD
|
dystrophin
|
|
chr12_-_51611476
|
0.165
|
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr6_-_47254143
|
0.161
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr1_-_93342706
|
0.154
|
NM_001252271
|
FAM69A
|
family with sequence similarity 69, member A
|
|
chr3_+_142375738
|
0.148
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr18_+_29027731
|
0.147
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr6_+_14117486
|
0.142
|
NM_001251901
|
CD83
|
CD83 molecule
|
|
chr10_+_5005601
|
0.135
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr3_-_196911001
|
0.135
|
NM_001204387
NM_001204388
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr19_-_56249739
|
0.133
|
NM_176820
|
NLRP9
|
NLR family, pyrin domain containing 9
|
|
chr3_+_130569368
|
0.133
|
NM_001199180
NM_001199181
NM_001199182
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chrX_-_77914824
|
0.133
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr12_+_20963637
|
0.132
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr2_+_128403438
|
0.131
|
NM_001161415
NM_001161416
NM_001161417
NM_005291
|
GPR17
|
G protein-coupled receptor 17
|
|
chr7_+_39017608
|
0.126
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chrX_+_65382627
|
0.124
|
|
HEPH
|
hephaestin
|
|
chr8_-_15095791
|
0.123
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr6_-_155777036
|
0.122
|
NM_015718
|
NOX3
|
NADPH oxidase 3
|
|
chr10_+_5005453
|
0.119
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr8_+_61591358
|
0.118
|
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr1_+_26503973
|
0.110
|
NM_006314
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr13_+_78109808
|
0.110
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr5_+_68710938
|
0.110
|
NM_001038603
NM_001244734
|
MARVELD2
|
MARVEL domain containing 2
|
|
chrX_-_33357725
|
0.109
|
NM_000109
|
DMD
|
dystrophin
|
|
chr8_-_49833823
|
0.106
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr6_-_47253962
|
0.106
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr3_-_57326709
|
0.106
|
NM_001142733
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chr5_+_140254930
|
0.105
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr4_-_72649734
|
0.104
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr13_+_43355685
|
0.104
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr2_+_113816214
|
0.097
|
NM_173170
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr4_+_78432905
|
0.097
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr15_+_40509628
|
0.097
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr22_-_22863504
|
0.096
|
NM_080764
|
ZNF280B
|
zinc finger protein 280B
|
|
chr6_-_26108320
|
0.095
|
NM_005323
|
HIST1H1T
|
histone cluster 1, H1t
|
|
chr7_-_41742666
|
0.095
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chrX_+_91090459
|
0.094
|
NM_001168360
NM_001168361
NM_001168362
NM_001168363
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr4_-_46125929
|
0.091
|
NM_173536
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1
|
|
chr6_-_2840683
|
0.090
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr19_-_49658645
|
0.089
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chrX_-_32173585
|
0.088
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr12_+_25205488
|
0.086
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr10_-_5045967
|
0.084
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chrX_-_55057408
|
0.084
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr1_+_156254069
|
0.083
|
NM_032323
|
TMEM79
|
transmembrane protein 79
|
|
chr20_-_6104056
|
0.082
|
NM_017671
|
FERMT1
|
fermitin family member 1
|
|
chr6_-_47277646
|
0.082
|
NM_014452
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr14_-_95599794
|
0.082
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr12_-_8219001
|
0.081
|
|
C3AR1
|
complement component 3a receptor 1
|
|
chr2_-_142889269
|
0.079
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr20_+_62795826
|
0.077
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr11_-_83393436
|
0.076
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr12_-_46633551
|
0.075
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr1_-_247054277
|
0.074
|
|
AHCTF1
|
AT hook containing transcription factor 1
|
|
chr5_+_55149149
|
0.074
|
NM_001242636
NM_001242639
|
IL31RA
|
interleukin 31 receptor A
|
|
chr2_-_158731622
|
0.073
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr21_-_42219038
|
0.073
|
NM_001389
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr12_+_56324945
|
0.073
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr13_-_52980628
|
0.073
|
NM_018676
NM_199263
|
THSD1
|
thrombospondin, type I, domain containing 1
|
|
chr4_-_87028805
|
0.073
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr20_+_9198036
|
0.072
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr3_+_97868169
|
0.070
|
NM_001005514
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14
|
|
chr11_+_5509914
|
0.068
|
NM_001005163
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1
|
|
chr17_-_39023461
|
0.068
|
NM_000223
|
KRT12
|
keratin 12
|
|
chr3_+_189349215
|
0.067
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr1_+_196743929
|
0.067
|
NM_001166624
NM_021023
|
CFHR3
|
complement factor H-related 3
|
|
chr7_+_93535819
|
0.066
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr22_+_22735134
|
0.066
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr4_-_151770611
|
0.065
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr16_+_82068958
|
0.064
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr17_-_48945338
|
0.064
|
NM_001243877
|
TOB1
|
transducer of ERBB2, 1
|
|
chr1_+_18081807
|
0.064
|
NM_030812
|
ACTL8
|
actin-like 8
|
|
chr19_+_39899749
|
0.064
|
|
|
|
|
chr15_+_74422920
|
0.062
|
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr4_+_90033967
|
0.062
|
NM_145715
|
TIGD2
|
tigger transposable element derived 2
|
|
chr4_+_41614933
|
0.061
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_-_137475076
|
0.061
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr7_-_38313232
|
0.060
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr16_+_82068910
|
0.059
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr15_+_74422742
|
0.057
|
NM_020851
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr1_+_67773046
|
0.057
|
NM_001559
|
IL12RB2
|
interleukin 12 receptor, beta 2
|
|
chr7_-_112430408
|
0.055
|
NM_022484
|
TMEM168
|
transmembrane protein 168
|
|
chr8_-_59412695
|
0.054
|
NM_000780
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1
|
|
chr1_+_167298382
|
0.054
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr7_-_50628744
|
0.053
|
NM_000790
NM_001242886
NM_001242887
NM_001242888
NM_001242889
NM_001242890
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr17_-_49124128
|
0.053
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr11_+_58710721
|
0.053
|
NM_001220496
NM_080661
|
GLYATL1
|
glycine-N-acyltransferase-like 1
|
|
chr8_+_80523376
|
0.053
|
|
STMN2
|
stathmin-like 2
|
|
chr1_-_116383323
|
0.053
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr2_-_241080072
|
0.053
|
NM_148961
|
OTOS
|
otospiralin
|
|
chr1_+_196621007
|
0.053
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr10_-_17171789
|
0.052
|
NM_001081
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr8_+_32579350
|
0.052
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr3_-_123339178
|
0.050
|
|
MYLK
|
myosin light chain kinase
|
|
chr17_-_38956146
|
0.050
|
NM_181535
|
KRT28
|
keratin 28
|
|
chr6_+_100054649
|
0.050
|
NM_021620
|
PRDM13
|
PR domain containing 13
|
|
chr17_-_10741398
|
0.049
|
NM_001101387
|
PIRT
|
phosphoinositide-interacting regulator of transient receptor potential channels
|
|
chr14_+_39703119
|
0.049
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr12_-_11214892
|
0.049
|
NM_176887
|
TAS2R46
|
taste receptor, type 2, member 46
|
|
chrX_+_150884506
|
0.049
|
NM_033085
|
FATE1
|
fetal and adult testis expressed 1
|
|
chr17_-_38938785
|
0.048
|
NM_181537
|
KRT27
|
keratin 27
|
|
chr4_+_41614911
|
0.047
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_-_153273683
|
0.046
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr10_+_77542518
|
0.045
|
NM_032024
|
C10orf11
|
chromosome 10 open reading frame 11
|
|
chr12_-_71148063
|
0.045
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr1_+_31882411
|
0.044
|
NM_001199038
|
SERINC2
|
serine incorporator 2
|
|
chr10_-_128209996
|
0.044
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr8_+_1993157
|
0.043
|
NM_003970
|
MYOM2
|
myomesin (M-protein) 2, 165kDa
|
|
chr3_-_190167597
|
0.043
|
NM_207316
|
TMEM207
|
transmembrane protein 207
|
|
chr9_-_125941304
|
0.043
|
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr3_-_13057130
|
0.043
|
|
|
|
|
chr10_+_29577989
|
0.043
|
NM_032517
|
LYZL1
|
lysozyme-like 1
|
|
chr1_+_81771844
|
0.043
|
|
LPHN2
|
latrophilin 2
|
|
chr5_+_55147206
|
0.043
|
NM_001242637
NM_001242638
NM_139017
|
IL31RA
|
interleukin 31 receptor A
|
|
chr6_+_127898318
|
0.042
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr8_-_101315457
|
0.042
|
NM_015435
|
RNF19A
|
ring finger protein 19A
|
|
chr6_+_88182624
|
0.041
|
NM_001168398
NM_006416
|
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1
|
|
chrX_-_15288588
|
0.041
|
NM_001168531
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr7_+_94297448
|
0.041
|
|
PEG10
|
paternally expressed 10
|
|
chr12_-_4488707
|
0.041
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr11_+_22696318
|
0.041
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr22_-_40289793
|
0.041
|
NM_152512
|
ENTHD1
|
ENTH domain containing 1
|
|
chr18_+_61144143
|
0.040
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr5_+_147443534
|
0.039
|
NM_001127698
NM_001127699
NM_006846
|
SPINK5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr13_+_109248499
|
0.039
|
NM_015011
|
MYO16
|
myosin XVI
|
|
chr9_+_35041101
|
0.039
|
NM_001040411
NM_001040412
|
C9orf131
|
chromosome 9 open reading frame 131
|
|
chr7_-_25268057
|
0.039
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr1_-_193155723
|
0.038
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr8_-_93075190
|
0.038
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_+_19290748
|
0.038
|
NM_017925
|
DENND4C
|
DENN/MADD domain containing 4C
|
|
chr1_+_167189948
|
0.038
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr1_-_39407449
|
0.038
|
NM_017821
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila)
|
|
chr4_+_166794409
|
0.037
|
NM_001204760
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr14_-_57960432
|
0.037
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr3_-_78666943
|
0.037
|
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr10_-_11574273
|
0.036
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr13_-_103719195
|
0.036
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr5_-_156390206
|
0.036
|
NM_001146726
NM_138379
|
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4
|
|
chr9_+_21409132
|
0.036
|
NM_002170
|
IFNA8
|
interferon, alpha 8
|
|
chr2_+_131278666
|
0.036
|
NM_032545
NM_001079530
|
CFC1
CFC1B
|
cripto, FRL-1, cryptic family 1
cripto, FRL-1, cryptic family 1B
|
|
chr7_-_83278478
|
0.035
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr13_+_109281569
|
0.035
|
NM_001198950
|
MYO16
|
myosin XVI
|
|
chr8_-_67090690
|
0.034
|
NM_000756
|
CRH
|
corticotropin releasing hormone
|
|
chr17_-_29641087
|
0.034
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr17_-_64225496
|
0.034
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr1_-_160617042
|
0.033
|
NM_003037
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr11_-_58980321
|
0.033
|
NM_001039396
|
MPEG1
|
macrophage expressed 1
|
|
chrX_-_15288172
|
0.033
|
NM_001031739
NM_001168530
NM_024087
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr1_+_84768333
|
0.033
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr3_+_111451326
|
0.033
|
NM_001134437
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr11_+_22689653
|
0.032
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr4_-_139163222
|
0.032
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr1_+_160709038
|
0.031
|
NM_021181
|
SLAMF7
|
SLAM family member 7
|
|
chr19_+_58180302
|
0.031
|
NM_152677
|
ZSCAN4
|
zinc finger and SCAN domain containing 4
|
|
chr2_-_131356859
|
0.031
|
NM_032545
NM_001079530
|
CFC1
CFC1B
|
cripto, FRL-1, cryptic family 1
cripto, FRL-1, cryptic family 1B
|
|
chr12_-_10562744
|
0.031
|
NM_001199805
|
KLRC4-KLRK1
|
KLRC4-KLRK1 readthrough
|
|
chr1_-_76397975
|
0.030
|
NM_080868
|
ASB17
|
ankyrin repeat and SOCS box containing 17
|
|
chr12_-_48963802
|
0.030
|
NM_002289
|
LALBA
|
lactalbumin, alpha-
|
|
chr22_+_22758538
|
0.030
|
|
|
|
|
chr7_+_143412938
|
0.030
|
NM_001130026
|
FAM115C
|
family with sequence similarity 115, member C
|
|
chr1_+_178694273
|
0.030
|
NM_152663
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr4_-_76944585
|
0.030
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr4_+_41614725
|
0.030
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chrX_+_123234462
|
0.030
|
|
STAG2
|
stromal antigen 2
|
|
chr2_+_128293312
|
0.029
|
NM_001080527
|
MYO7B
|
myosin VIIB
|
|
chr8_-_124749573
|
0.029
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chr4_+_95376396
|
0.029
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr2_+_27799388
|
0.029
|
NM_032266
|
C2orf16
|
chromosome 2 open reading frame 16
|
|
chr16_+_81272293
|
0.029
|
NM_017429
|
BCMO1
|
beta-carotene 15,15'-monooxygenase 1
|
|
chr2_+_183582667
|
0.029
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr12_-_71148331
|
0.028
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr21_-_39956823
|
0.028
|
NM_001243429
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr7_-_92855780
|
0.028
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|