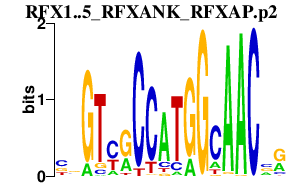
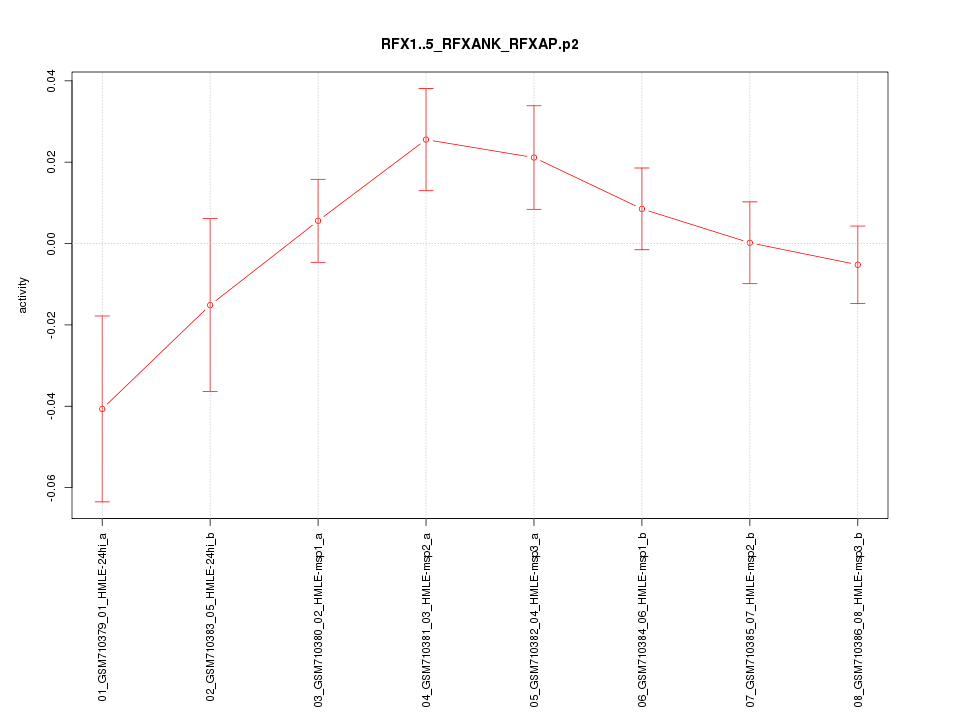
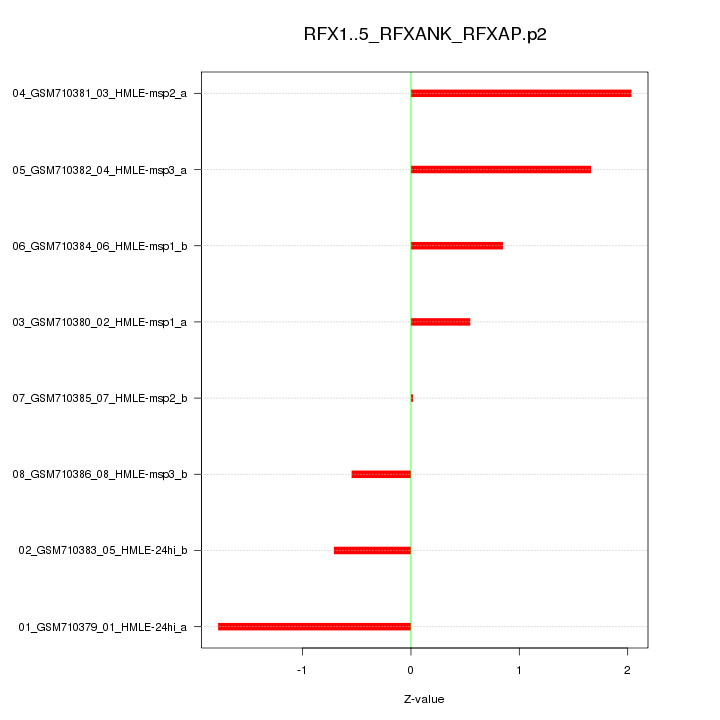
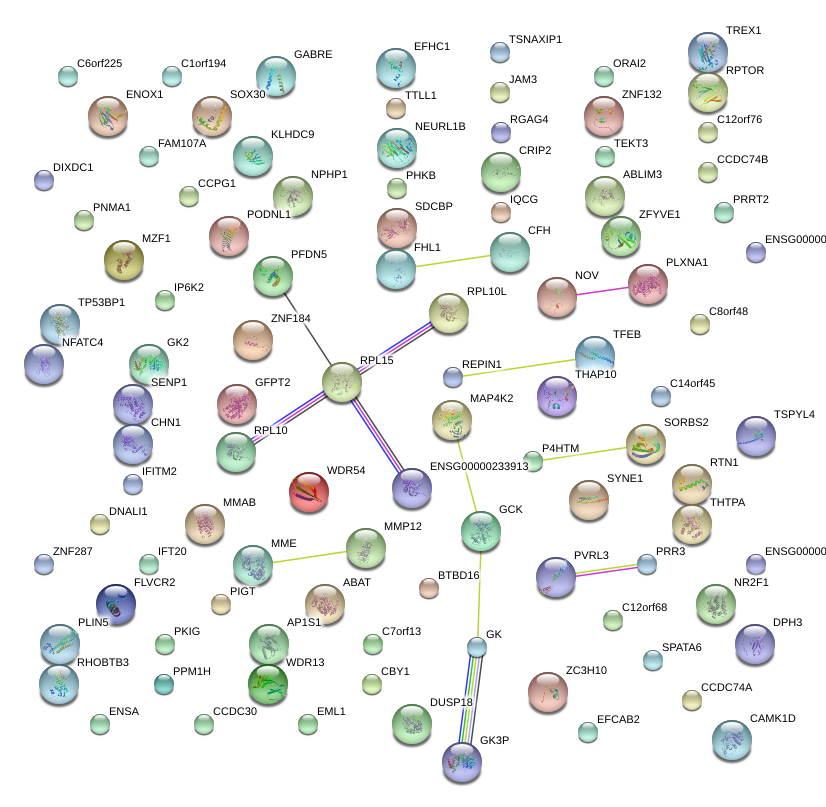
|
chr5_-_179780314
|
1.589
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr17_-_5404304
|
1.532
|
NM_001162371
|
LOC728392
|
uncharacterized LOC728392
|
|
chr14_-_60337350
|
1.396
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr14_+_105941061
|
1.349
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr5_+_92918924
|
1.156
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr11_+_133938819
|
0.956
|
NM_001205329
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr3_-_197676755
|
0.923
|
NM_001134435
|
IQCG
|
IQ motif containing G
|
|
chr14_+_100259445
|
0.918
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr1_+_42928873
|
0.901
|
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr11_+_133938965
|
0.820
|
|
JAM3
|
junctional adhesion molecule 3
|
|
chr8_+_120428551
|
0.819
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr6_+_30524662
|
0.808
|
|
PRR3
|
proline rich 3
|
|
chr2_-_175869910
|
0.800
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr16_+_29823557
|
0.795
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr1_-_109656478
|
0.779
|
NM_001122961
|
C1orf194
|
chromosome 1 open reading frame 194
|
|
chr22_-_31063786
|
0.777
|
NM_152511
|
DUSP18
|
dual specificity phosphatase 18
|
|
chr22_-_43485333
|
0.767
|
NM_012263
|
TTLL1
|
tubulin tyrosine ligase-like family, member 1
|
|
chr1_-_48937791
|
0.745
|
NM_019073
|
SPATA6
|
spermatogenesis associated 6
|
|
chr3_-_196439067
|
0.737
|
NM_032898
|
CEP19
|
centrosomal protein 19kDa
|
|
chr12_-_63328663
|
0.719
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr16_+_8806825
|
0.714
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr3_-_58563070
|
0.712
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr1_+_161068150
|
0.702
|
NM_001007255
NM_152366
|
KLHDC9
|
kelch domain containing 9
|
|
chr1_+_245133283
|
0.694
|
|
EFCAB2
|
EF-hand calcium binding domain 2
|
|
chr3_+_154797435
|
0.662
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr20_+_43160435
|
0.634
|
NM_007066
NM_181804
NM_181805
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr6_+_112408673
|
0.632
|
NM_001033564
|
C6orf225
|
chromosome 6 open reading frame 225
|
|
chr7_+_102073953
|
0.618
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr17_-_64187814
|
0.616
|
NM_145036
NM_001199165
|
CEP112
|
centrosomal protein 112kDa
|
|
chr19_-_58951588
|
0.608
|
NM_003433
|
ZNF132
|
zinc finger protein 132
|
|
chr17_-_16472463
|
0.604
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chr7_+_102074002
|
0.596
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr14_+_76044939
|
0.595
|
NM_017791
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr6_+_30525050
|
0.585
|
|
PRR3
|
proline rich 3
|
|
chrX_+_48456552
|
0.564
|
|
WDR13
|
WD repeat domain 13
|
|
chr10_+_12391471
|
0.563
|
NM_020397
NM_153498
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr17_+_78518742
|
0.561
|
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr19_-_59084735
|
0.559
|
|
MZF1
|
myeloid zinc finger 1
|
|
chr8_+_13424351
|
0.559
|
NM_001007090
|
C8orf48
|
chromosome 8 open reading frame 48
|
|
chr3_+_49027770
|
0.557
|
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chrX_+_48456122
|
0.556
|
|
WDR13
|
WD repeat domain 13
|
|
chr14_-_73453651
|
0.553
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr21_+_46707949
|
0.536
|
|
LOC642852
|
uncharacterized LOC642852
|
|
chr3_+_183165402
|
0.535
|
|
LOC100505687
|
uncharacterized LOC100505687
|
|
chr15_+_55700722
|
0.533
|
NM_001198784
|
FLJ27352
|
uncharacterized LOC145788
|
|
chr14_+_74486049
|
0.525
|
NM_025057
|
C14orf45
|
chromosome 14 open reading frame 45
|
|
chrX_+_48456220
|
0.523
|
NM_001166426
|
WDR13
|
WD repeat domain 13
|
|
chr1_-_48937676
|
0.520
|
|
SPATA6
|
spermatogenesis associated 6
|
|
chr3_-_16306283
|
0.507
|
NM_001047434
NM_206831
|
DPH3
|
DPH3, KTI11 homolog (S. cerevisiae)
|
|
chrX_-_71351750
|
0.506
|
NM_001024455
|
RGAG4
|
retrotransposon gag domain containing 4
|
|
chr7_-_44228880
|
0.501
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr3_+_49027281
|
0.498
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr13_-_44361032
|
0.492
|
NM_001242863
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr5_+_172068188
|
0.491
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr19_-_59084630
|
0.491
|
|
MZF1
|
myeloid zinc finger 1
|
|
chr16_+_29823408
|
0.485
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr6_+_30524485
|
0.476
|
NM_001077497
NM_025263
|
PRR3
|
proline rich 3
|
|
chr1_+_38022514
|
0.474
|
NM_003462
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr2_+_242811880
|
0.474
|
NM_173821
|
CXXC11
|
CXXC finger protein 11
|
|
chrX_+_135251795
|
0.472
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr5_-_157098444
|
0.469
|
|
SOX30
|
SRY (sex determining region Y)-box 30
|
|
chr2_+_74648856
|
0.459
|
NM_032118
|
WDR54
|
WD repeat domain 54
|
|
chr12_+_48577365
|
0.456
|
NM_001013635
|
C12orf68
|
chromosome 12 open reading frame 68
|
|
chr7_+_150065968
|
0.456
|
|
REPIN1
|
replication initiator 1
|
|
chr17_-_26662439
|
0.438
|
NM_174887
|
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas)
|
|
chr14_+_24025188
|
0.437
|
NM_001126339
NM_024328
|
THTPA
|
thiamine triphosphatase
|
|
chr22_+_27053392
|
0.435
|
|
MIAT
|
myocardial infarction associated transcript (non-protein coding)
|
|
chr6_-_152702329
|
0.434
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr7_+_100797685
|
0.432
|
NM_001283
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit
|
|
chr10_+_124030820
|
0.429
|
NM_144587
|
BTBD16
|
BTB (POZ) domain containing 16
|
|
chrX_-_151143096
|
0.426
|
NM_004961
|
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon
|
|
chrX_+_48455875
|
0.418
|
NM_017883
|
WDR13
|
WD repeat domain 13
|
|
chr12_+_56511969
|
0.418
|
NM_032786
|
ZC3H10
|
zinc finger CCCH-type containing 10
|
|
chrX_+_135251454
|
0.414
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr5_+_95066628
|
0.411
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr3_+_48507209
|
0.410
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr19_-_59084928
|
0.409
|
NM_003422
NM_198055
|
MZF1
|
myeloid zinc finger 1
|
|
chr14_-_74180341
|
0.409
|
NM_006029
|
PNMA1
|
paraneoplastic antigen MA1
|
|
chr12_+_53689234
|
0.409
|
NM_002624
NM_145897
|
PFDN5
|
prefoldin subunit 5
|
|
chr2_-_130902568
|
0.408
|
NM_207310
|
CCDC74B
|
coiled-coil domain containing 74B
|
|
chr15_-_55700456
|
0.408
|
NM_004748
NM_001204450
NM_001204451
NM_020739
|
CCPG1
|
cell cycle progression 1
|
|
chr19_-_4535202
|
0.406
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr7_+_150065330
|
0.401
|
|
REPIN1
|
replication initiator 1
|
|
chr8_+_59465783
|
0.400
|
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr19_+_18530423
|
0.397
|
|
|
|
|
chr3_-_48754589
|
0.394
|
NM_001005909
NM_001005910
NM_001005911
NM_001146178
NM_001146179
NM_001190316
NM_001190317
NM_016291
|
IP6K2
|
inositol hexakisphosphate kinase 2
|
|
chr14_+_24838301
|
0.393
|
NM_001198966
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr16_+_47495166
|
0.393
|
NM_000293
NM_001031835
|
PHKB
|
phosphorylase kinase, beta
|
|
chr6_-_27440459
|
0.392
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr12_-_110011286
|
0.390
|
NM_052845
|
MMAB
|
methylmalonic aciduria (cobalamin deficiency) cblB type
|
|
chrX_+_30671602
|
0.389
|
|
GK
|
glycerol kinase
|
|
chr7_-_156433115
|
0.388
|
|
C7orf13
|
chromosome 7 open reading frame 13
|
|
chr7_+_150065859
|
0.383
|
NM_001099695
NM_001099696
NM_013400
|
REPIN1
|
replication initiator 1
|
|
chr12_-_48499840
|
0.381
|
|
SENP1
|
SUMO1/sentrin specific peptidase 1
|
|
chr15_-_43802666
|
0.380
|
NM_005657
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr1_-_150601554
|
0.378
|
NM_207045
NM_207046
NM_207047
|
ENSA
|
endosulfine alpha
|
|
chr2_+_132285469
|
0.373
|
NM_138770
|
CCDC74A
|
coiled-coil domain containing 74A
|
|
chr16_+_67840976
|
0.372
|
NM_018430
|
TSNAXIP1
|
translin-associated factor X interacting protein 1
|
|
chr12_-_110486275
|
0.370
|
|
C12orf76
|
chromosome 12 open reading frame 76
|
|
chr16_+_8814567
|
0.369
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr19_-_14048877
|
0.369
|
|
PODNL1
|
podocan-like 1
|
|
chr15_-_71184611
|
0.369
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr5_+_148520985
|
0.366
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr20_+_44044706
|
0.365
|
NM_001184728
NM_001184729
NM_001184730
NM_015937
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T
|
|
chr6_+_52284948
|
0.360
|
NM_018100
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chrX_+_48455801
|
0.359
|
|
WDR13
|
WD repeat domain 13
|
|
chr22_+_39052671
|
0.359
|
|
CBY1
|
chibby homolog 1 (Drosophila)
|
|
chrX_+_30671537
|
0.357
|
|
GK
|
glycerol kinase
|
|
chr12_+_53689350
|
0.354
|
|
PFDN5
|
prefoldin subunit 5
|
|
chr6_-_43477980
|
0.351
|
|
LRRC73
|
leucine rich repeat containing 73
|
|
chr22_+_39052628
|
0.347
|
NM_001002880
NM_015373
|
CBY1
|
chibby homolog 1 (Drosophila)
|
|
chr17_-_15244898
|
0.346
|
NM_031898
|
TEKT3
|
tektin 3
|
|
chr6_-_41703938
|
0.346
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr6_-_116575170
|
0.346
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr2_-_110962577
|
0.342
|
NM_000272
NM_001128178
NM_001128179
NM_207181
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chrX_+_48455915
|
0.342
|
|
WDR13
|
WD repeat domain 13
|
|
chr4_-_186733372
|
0.342
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_+_111808017
|
0.339
|
|
DIXDC1
|
DIX domain containing 1
|
|
chr5_-_133702764
|
0.337
|
NM_001113575
NM_016508
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr4_+_153701113
|
0.335
|
|
ARFIP1
|
ADP-ribosylation factor interacting protein 1
|
|
chr11_-_66360485
|
0.334
|
NM_018219
|
CCDC87
|
coiled-coil domain containing 87
|
|
chr16_-_69760367
|
0.334
|
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr1_+_155147051
|
0.334
|
|
TRIM46
|
tripartite motif containing 46
|
|
chr19_-_50528683
|
0.334
|
NM_001025778
NM_016440
|
VRK3
|
vaccinia related kinase 3
|
|
chr1_-_1551041
|
0.330
|
|
MIB2
|
mindbomb homolog 2 (Drosophila)
|
|
chr16_+_47495225
|
0.330
|
|
PHKB
|
phosphorylase kinase, beta
|
|
chr7_-_14030864
|
0.330
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr1_+_3541550
|
0.330
|
NM_182752
|
TPRG1L
|
tumor protein p63 regulated 1-like
|
|
chr21_+_38445562
|
0.327
|
NM_001001894
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr6_-_152702450
|
0.323
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr17_-_80170656
|
0.323
|
NM_198082
|
CCDC57
|
coiled-coil domain containing 57
|
|
chr19_-_50528627
|
0.323
|
|
VRK3
|
vaccinia related kinase 3
|
|
chr11_+_46958228
|
0.323
|
NM_001003676
NM_001003677
NM_001003678
NM_024113
|
C11orf49
|
chromosome 11 open reading frame 49
|
|
chr9_-_131418931
|
0.322
|
|
WDR34
|
WD repeat domain 34
|
|
chr12_-_54653369
|
0.320
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr3_+_9851643
|
0.318
|
NM_001025930
|
TTLL3
|
tubulin tyrosine ligase-like family, member 3
|
|
chr12_-_63328857
|
0.318
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr1_-_163113938
|
0.317
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr3_+_184056304
|
0.315
|
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr17_+_19281726
|
0.315
|
NM_002749
NM_139034
|
MAPK7
|
mitogen-activated protein kinase 7
|
|
chr16_+_3550916
|
0.315
|
NM_015041
|
CLUAP1
|
clusterin associated protein 1
|
|
chr16_+_67312688
|
0.314
|
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr9_-_131419004
|
0.310
|
NM_052844
|
WDR34
|
WD repeat domain 34
|
|
chr16_+_57278979
|
0.308
|
NM_012106
|
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein
|
|
chr8_+_16884745
|
0.307
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr2_+_39103102
|
0.306
|
NM_001145450
|
MORN2
|
MORN repeat containing 2
|
|
chr4_-_8430185
|
0.303
|
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr20_-_50179338
|
0.303
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr16_-_69760354
|
0.302
|
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr6_+_35226685
|
0.300
|
|
ZNF76
|
zinc finger protein 76
|
|
chr18_+_51795848
|
0.300
|
NM_007195
|
POLI
|
polymerase (DNA directed) iota
|
|
chr11_+_66278076
|
0.300
|
NM_024649
|
BBS1
|
Bardet-Biedl syndrome 1
|
|
chr7_-_14031049
|
0.299
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr21_+_40817793
|
0.296
|
NM_001001713
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein
|
|
chr19_+_10527448
|
0.293
|
NM_001243121
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr12_+_51818669
|
0.290
|
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr19_+_35630391
|
0.287
|
NM_005031
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr5_-_150537261
|
0.287
|
|
ANXA6
|
annexin A6
|
|
chr17_+_260092
|
0.286
|
NM_001013672
|
C17orf97
|
chromosome 17 open reading frame 97
|
|
chr4_+_123653856
|
0.283
|
NM_001178007
NM_152618
|
BBS12
|
Bardet-Biedl syndrome 12
|
|
chr12_+_51818592
|
0.282
|
NM_001039960
NM_004858
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr12_-_131323616
|
0.281
|
NM_001980
NM_194356
|
STX2
|
syntaxin 2
|
|
chr2_+_237994500
|
0.279
|
|
COPS8
|
COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis)
|
|
chr9_+_35829208
|
0.279
|
NM_016446
NM_001042589
|
TMEM8B
|
transmembrane protein 8B
|
|
chr6_+_35227478
|
0.273
|
NM_003427
|
ZNF76
|
zinc finger protein 76
|
|
chr14_-_20774089
|
0.273
|
|
TTC5
|
tetratricopeptide repeat domain 5
|
|
chr17_-_53045972
|
0.272
|
NM_001162861
NM_001162862
NM_004375
|
COX11
|
COX11 cytochrome c oxidase assembly homolog (yeast)
|
|
chr9_+_100069909
|
0.269
|
NM_020893
|
C9orf174
|
chromosome 9 open reading frame 174
|
|
chr11_+_66360664
|
0.268
|
NM_005125
|
CCS
|
copper chaperone for superoxide dismutase
|
|
chr16_-_69760444
|
0.266
|
NM_000903
NM_001025433
NM_001025434
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr16_+_72698996
|
0.264
|
|
|
|
|
chr2_+_48667907
|
0.263
|
NM_001135629
NM_001193475
NM_152994
|
PPP1R21
|
protein phosphatase 1, regulatory subunit 21
|
|
chr22_+_24309025
|
0.262
|
NM_001084393
|
DDTL
|
D-dopachrome tautomerase-like
|
|
chr19_-_10679328
|
0.261
|
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr13_-_96294282
|
0.260
|
|
DZIP1
|
DAZ interacting protein 1
|
|
chr11_+_63706433
|
0.260
|
NM_024771
|
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae)
|
|
chr6_+_35227234
|
0.260
|
|
ZNF76
|
zinc finger protein 76
|
|
chr19_+_49977817
|
0.259
|
NM_001204503
|
FLT3LG
|
fms-related tyrosine kinase 3 ligand
|
|
chr15_-_43785368
|
0.259
|
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr1_+_245133067
|
0.259
|
NM_032328
|
EFCAB2
|
EF-hand calcium binding domain 2
|
|
chr19_-_58609606
|
0.258
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr11_+_111807857
|
0.258
|
NM_001037954
|
DIXDC1
|
DIX domain containing 1
|
|
chr19_-_18392436
|
0.256
|
|
JUND
|
jun D proto-oncogene
|
|
chr1_-_36915969
|
0.256
|
NM_145047
NM_206837
|
OSCP1
|
organic solute carrier partner 1
|
|
chr17_-_1012257
|
0.256
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr8_+_59465818
|
0.256
|
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr14_-_20774129
|
0.256
|
NM_138376
|
TTC5
|
tetratricopeptide repeat domain 5
|
|
chr12_-_4754275
|
0.255
|
NM_006422
|
AKAP3
|
A kinase (PRKA) anchor protein 3
|
|
chr1_+_78444922
|
0.255
|
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr9_-_34620460
|
0.254
|
|
DCTN3
|
dynactin 3 (p22)
|
|
chr9_-_34620483
|
0.253
|
|
DCTN3
|
dynactin 3 (p22)
|
|
chr7_-_100964923
|
0.251
|
NM_001130820
NM_001130821
NM_001130822
NM_022777
|
RABL5
|
RAB, member RAS oncogene family-like 5
|
|
chr19_+_50879681
|
0.248
|
NM_007121
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chr19_+_46850245
|
0.248
|
NM_001204284
NM_006247
|
PPP5C
|
protein phosphatase 5, catalytic subunit
|
|
chr6_-_84937313
|
0.248
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr22_-_44893903
|
0.248
|
NM_032287
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr3_-_196045037
|
0.247
|
NM_152773
|
TCTEX1D2
|
Tctex1 domain containing 2
|
|
chr9_+_127615695
|
0.245
|
NM_001045476
|
WDR38
|
WD repeat domain 38
|
|
chr19_-_18392413
|
0.244
|
NM_005354
|
JUND
|
jun D proto-oncogene
|
|
chr9_-_34620489
|
0.243
|
NM_007234
NM_024348
|
DCTN3
|
dynactin 3 (p22)
|