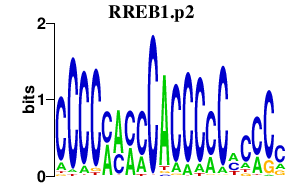
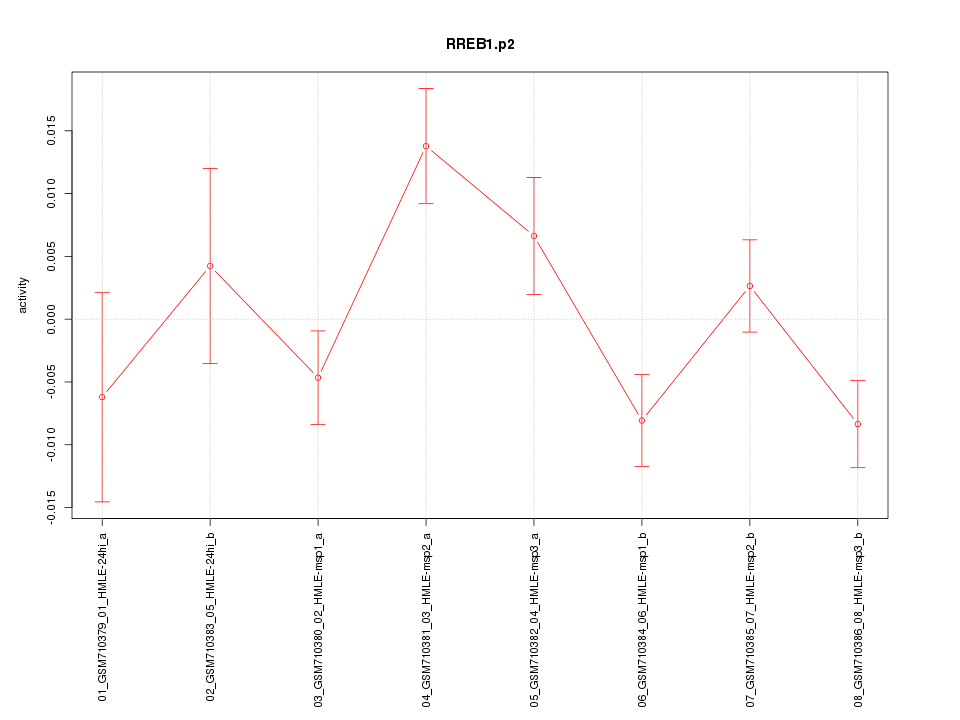
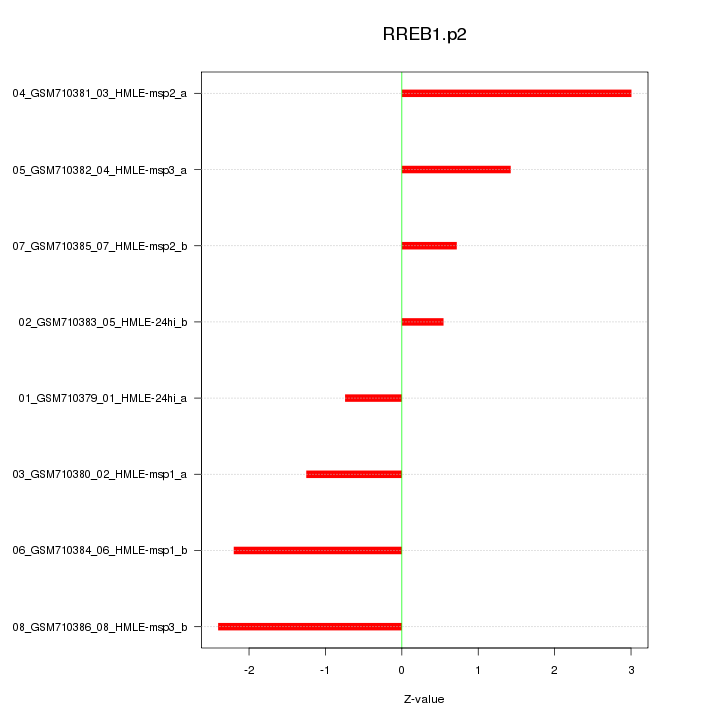
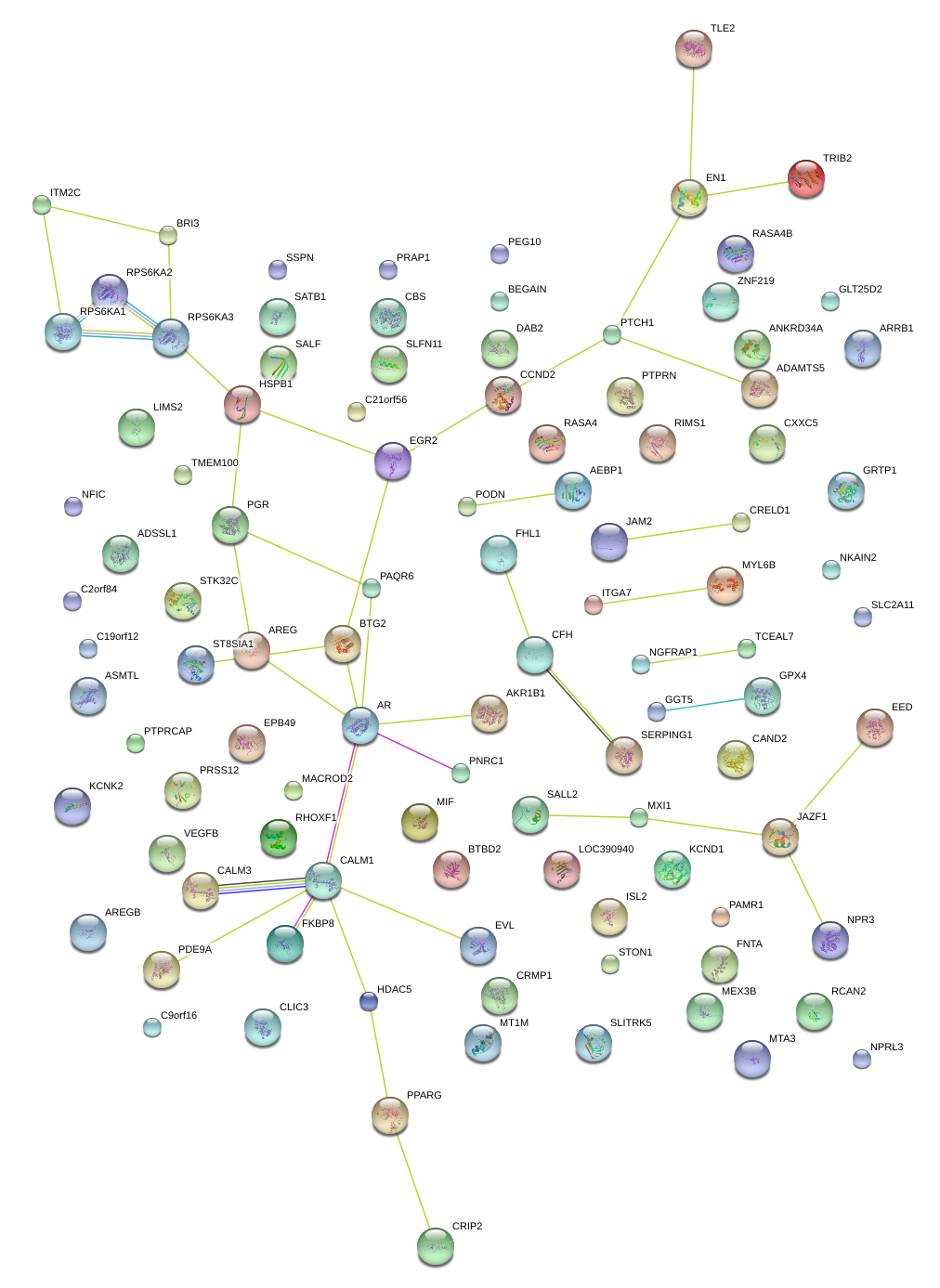
|
chr13_-_114018201
|
1.227
|
|
GRTP1
|
growth hormone regulated TBC protein 1
|
|
chr6_-_46459010
|
1.019
|
NM_001251974
|
RCAN2
|
regulator of calcineurin 2
|
|
chr21_-_44495894
|
0.939
|
NM_000071
|
CBS
|
cystathionine-beta-synthase
|
|
chr21_+_27011766
|
0.931
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr13_-_114018365
|
0.908
|
NM_024719
|
GRTP1
|
growth hormone regulated TBC protein 1
|
|
chr21_+_27012067
|
0.796
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr10_+_111969988
|
0.784
|
NM_001008541
|
MXI1
|
MAX interactor 1
|
|
chr19_+_3366575
|
0.704
|
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr1_-_184006491
|
0.693
|
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr9_+_130922471
|
0.641
|
NM_024112
|
C9orf16
|
chromosome 9 open reading frame 16
|
|
chrX_+_66763868
|
0.639
|
NM_000044
|
AR
|
androgen receptor
|
|
chrX_+_102585113
|
0.616
|
NM_152278
|
TCEAL7
|
transcription elongation factor A (SII)-like 7
|
|
chr21_+_44073650
|
0.611
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr21_-_44496423
|
0.606
|
NM_001178008
NM_001178009
|
CBS
|
cystathionine-beta-synthase
|
|
chr21_+_27011588
|
0.596
|
NM_021219
|
JAM2
|
junctional adhesion molecule 2
|
|
chr11_-_2019007
|
0.550
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr21_-_47604300
|
0.530
|
|
C21orf56
|
chromosome 21 open reading frame 56
|
|
chr21_-_47604360
|
0.529
|
NM_001142854
NM_032261
|
C21orf56
|
chromosome 21 open reading frame 56
|
|
chr21_-_47604340
|
0.526
|
|
C21orf56
|
chromosome 21 open reading frame 56
|
|
chr11_-_2018703
|
0.510
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chrX_-_119249774
|
0.503
|
NM_139282
|
RHOXF1
|
Rhox homeobox family, member 1
|
|
chr1_-_156217717
|
0.490
|
NM_198406
NM_024897
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr12_-_22487566
|
0.490
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr21_+_27012045
|
0.488
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr6_-_167275657
|
0.462
|
NM_001006932
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chrX_+_102632108
|
0.459
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr2_-_128432642
|
0.459
|
NM_001136037
NM_001161403
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr19_-_18653680
|
0.455
|
|
FKBP8
|
FK506 binding protein 8, 38kDa
|
|
chr17_-_42201009
|
0.445
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr12_+_26348268
|
0.438
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr21_-_28339405
|
0.428
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr2_+_42795844
|
0.417
|
|
MTA3
|
metastasis associated 1 family, member 3
|
|
chr14_-_21566497
|
0.415
|
|
ZNF219
|
zinc finger protein 219
|
|
chr10_+_135160843
|
0.413
|
NM_001145201
NM_145202
|
PRAP1
|
proline-rich acidic protein 1
|
|
chr11_-_2018568
|
0.410
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr11_+_64001974
|
0.402
|
NM_001243733
NM_003377
|
VEGFB
|
vascular endothelial growth factor B
|
|
chr11_-_75062650
|
0.402
|
NM_004041
NM_020251
|
ARRB1
|
arrestin, beta 1
|
|
chrX_-_1572648
|
0.398
|
NM_001173473
|
ASMTL
|
acetylserotonin O-methyltransferase-like
|
|
chr14_-_21994342
|
0.396
|
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr5_+_32711437
|
0.396
|
NM_000908
NM_001204375
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr1_+_145472454
|
0.394
|
|
ANKRD34A
|
ankyrin repeat domain 34A
|
|
chrX_+_135229536
|
0.387
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_+_72596699
|
0.384
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr19_+_1104007
|
0.376
|
|
GPX4
|
glutathione peroxidase 4 (phospholipid hydroperoxidase)
|
|
chr19_-_30205962
|
0.371
|
NM_001031726
|
C19orf12
|
chromosome 19 open reading frame 12
|
|
chrX_+_102631408
|
0.371
|
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr12_+_26348488
|
0.363
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr22_+_24199058
|
0.363
|
NM_001024938
NM_030807
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11
|
|
chr7_+_75932992
|
0.356
|
|
HSPB1
|
heat shock 27kDa protein 1
|
|
chr1_+_1076275
|
0.351
|
|
LOC254099
|
uncharacterized LOC254099
|
|
chr7_+_75932865
|
0.351
|
|
HSPB1
|
heat shock 27kDa protein 1
|
|
chr7_+_44144002
|
0.350
|
|
AEBP1
|
AE binding protein 1
|
|
chr19_+_3366589
|
0.349
|
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chrY_-_1522648
|
0.345
|
NM_001173473
|
ASMTL
|
acetylserotonin O-methyltransferase-like
|
|
chr19_+_3366536
|
0.345
|
NM_001245002
NM_001245004
NM_005597
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr12_-_56101465
|
0.338
|
|
ITGA7
|
integrin, alpha 7
|
|
chr19_+_44084695
|
0.335
|
NM_001193622
|
LOC390940
|
uncharacterized protein ENSP00000244321
|
|
chr9_-_139890984
|
0.334
|
NM_004669
|
CLIC3
|
chloride intracellular channel 3
|
|
chr12_+_56546203
|
0.332
|
NM_001199629
NM_002475
|
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle
|
|
chr7_+_94285736
|
0.332
|
|
PEG10
|
paternally expressed 10
|
|
chr11_-_35547144
|
0.331
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr7_+_94285676
|
0.330
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr22_-_24640850
|
0.328
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr7_+_94285620
|
0.327
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr13_+_88324777
|
0.327
|
NM_015567
|
SLITRK5
|
SLIT and NTRK-like family, member 5
|
|
chr11_+_57365026
|
0.326
|
NM_000062
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr7_+_44143945
|
0.326
|
NM_001129
|
AEBP1
|
AE binding protein 1
|
|
chr10_-_64578926
|
0.324
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr19_+_44084758
|
0.321
|
|
LOC390940
|
uncharacterized protein ENSP00000244321
|
|
chr17_-_33700619
|
0.320
|
NM_001104587
NM_001104588
NM_001104589
NM_001104590
NM_152270
|
SLFN11
|
schlafen family member 11
|
|
chr14_+_100438095
|
0.319
|
|
EVL
|
Enah/Vasp-like
|
|
chr15_-_82338348
|
0.317
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr6_-_167275997
|
0.316
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr1_+_203274646
|
0.315
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr3_+_12838170
|
0.314
|
NM_001162499
NM_012298
|
CAND2
|
cullin-associated and neddylation-dissociated 2 (putative)
|
|
chr14_+_105941061
|
0.313
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr7_-_28220342
|
0.312
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr3_-_18466759
|
0.310
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr11_-_1769086
|
0.306
|
|
IFITM10
|
interferon induced transmembrane protein 10
|
|
chr17_-_42200957
|
0.306
|
|
HDAC5
|
histone deacetylase 5
|
|
chr15_+_76629101
|
0.304
|
NM_145805
|
ISL2
|
ISL LIM homeobox 2
|
|
chr2_+_48757292
|
0.298
|
NM_001198595
NM_006873
|
STON1
|
stonin 1
|
|
chr19_-_18654280
|
0.295
|
|
FKBP8
|
FK506 binding protein 8, 38kDa
|
|
chr2_+_12856813
|
0.294
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr3_+_9975513
|
0.289
|
NM_015513
NM_001031717
NM_001077415
|
CRELD1
|
cysteine-rich with EGF-like domains 1
|
|
chr1_+_53527723
|
0.288
|
NM_001199080
NM_001199082
NM_153703
|
PODN
|
podocan
|
|
chr4_-_5890266
|
0.286
|
NM_001313
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr19_+_1104068
|
0.286
|
|
GPX4
|
glutathione peroxidase 4 (phospholipid hydroperoxidase)
|
|
chr6_+_72596405
|
0.286
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr19_+_1104614
|
0.284
|
NM_001039848
|
GPX4
|
glutathione peroxidase 4 (phospholipid hydroperoxidase)
|
|
chr19_-_3028901
|
0.283
|
NM_003260
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr9_-_98270321
|
0.283
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr14_+_105196225
|
0.282
|
NM_199165
|
ADSSL1
|
adenylosuccinate synthase like 1
|
|
chr6_+_89791510
|
0.279
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr5_-_39424930
|
0.278
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr19_+_1103935
|
0.277
|
NM_001039847
NM_002085
|
GPX4
|
glutathione peroxidase 4 (phospholipid hydroperoxidase)
|
|
chr6_+_124125068
|
0.276
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr16_+_56666533
|
0.275
|
NM_176870
|
MT1M
|
metallothionein 1M
|
|
chr11_-_2019070
|
0.271
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr19_+_47105327
|
0.267
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr5_+_139027907
|
0.265
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr17_-_53809444
|
0.265
|
NM_001099640
|
TMEM100
|
transmembrane protein 100
|
|
chrX_+_135230736
|
0.262
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr2_+_24299751
|
0.259
|
|
LOC375190
|
UPF0638 protein B
|
|
chr1_+_215256559
|
0.254
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr4_-_5890142
|
0.254
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr11_+_57365577
|
0.254
|
NM_001032295
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr19_-_2015628
|
0.250
|
NM_017797
|
BTBD2
|
BTB (POZ) domain containing 2
|
|
chr12_+_4382882
|
0.248
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chrX_-_48828250
|
0.248
|
NM_004979
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1
|
|
chr3_+_12329332
|
0.246
|
NM_005037
NM_138712
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr14_-_101034261
|
0.246
|
NM_001159531
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr5_+_32711729
|
0.246
|
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr2_-_220173693
|
0.246
|
NM_001199764
|
PTPRN
|
protein tyrosine phosphatase, receptor type, N
|
|
chr7_-_102257199
|
0.244
|
NM_001079877
NM_006989
|
RASA4
|
RAS p21 protein activator 4
|
|
chr20_+_13976014
|
0.244
|
NM_080676
|
MACROD2
|
MACRO domain containing 2
|
|
chr2_+_42795601
|
0.243
|
NM_020744
|
MTA3
|
metastasis associated 1 family, member 3
|
|
chr2_-_119605758
|
0.240
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr19_-_3047632
|
0.240
|
NM_001144761
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr4_-_119273874
|
0.240
|
NM_003619
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin)
|
|
chr10_-_134145333
|
0.239
|
|
STK32C
|
serine/threonine kinase 32C
|
|
chr11_-_67205151
|
0.235
|
NM_005608
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein
|
|
chr7_+_97911023
|
0.234
|
|
BRI3
|
brain protein I3
|
|
chrX_+_103031433
|
0.234
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr9_-_21994329
|
0.233
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr19_-_11564568
|
0.232
|
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr12_-_53614121
|
0.231
|
NM_001042728
NM_001243732
|
RARG
|
retinoic acid receptor, gamma
|
|
chr3_-_21792815
|
0.231
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr1_-_47080793
|
0.231
|
NM_145279
|
MOB3C
|
MOB kinase activator 3C
|
|
chr9_-_21994414
|
0.228
|
NM_058195
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr14_+_96505566
|
0.228
|
NM_001252507
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr20_-_44144263
|
0.227
|
NM_006652
|
SPINT3
|
serine peptidase inhibitor, Kunitz type, 3
|
|
chr10_+_83635069
|
0.227
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr2_-_128433330
|
0.226
|
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr1_-_231004173
|
0.225
|
NM_032800
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr12_+_57482886
|
0.225
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr4_+_184827882
|
0.224
|
|
|
|
|
chr11_-_8739398
|
0.224
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr14_+_24837225
|
0.223
|
NM_001198965
NM_004554
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chrX_-_152486073
|
0.222
|
NM_004988
|
MAGEA1
|
melanoma antigen family A, 1 (directs expression of antigen MZ2-E)
|
|
chr19_+_35521591
|
0.222
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr2_+_30369749
|
0.222
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr1_-_231004050
|
0.221
|
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr16_-_4588379
|
0.221
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr12_+_70759974
|
0.220
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr1_-_1535446
|
0.220
|
NM_001242659
|
LOC643988
|
uncharacterized LOC643988
|
|
chr4_-_84030995
|
0.220
|
NM_001130716
|
PLAC8
|
placenta-specific 8
|
|
chr8_+_144635556
|
0.219
|
NM_001166237
|
GSDMD
|
gasdermin D
|
|
chr19_-_3062191
|
0.219
|
NM_001130
NM_198970
|
AES
|
amino-terminal enhancer of split
|
|
chr14_+_105190533
|
0.219
|
NM_152328
|
ADSSL1
|
adenylosuccinate synthase like 1
|
|
chr5_+_139027868
|
0.219
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr4_+_184827805
|
0.218
|
|
STOX2
|
storkhead box 2
|
|
chr16_+_68573660
|
0.217
|
NM_133458
|
ZFP90
|
zinc finger protein 90 homolog (mouse)
|
|
chrX_-_100872926
|
0.216
|
NM_001009584
NM_001184768
NM_019007
|
ARMCX6
|
armadillo repeat containing, X-linked 6
|
|
chr22_+_24200040
|
0.214
|
NM_001024939
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11
|
|
chr17_-_19290471
|
0.214
|
NM_001198695
NM_002404
|
MFAP4
|
microfibrillar-associated protein 4
|
|
chr3_-_114866090
|
0.214
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_-_66453561
|
0.214
|
NM_017983
|
WIPI1
|
WD repeat domain, phosphoinositide interacting 1
|
|
chr11_-_17498419
|
0.213
|
NM_000352
|
ABCC8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8
|
|
chr19_+_41082792
|
0.211
|
|
SHKBP1
|
SH3KBP1 binding protein 1
|
|
chr7_-_150974179
|
0.211
|
NM_003078
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr2_+_219264719
|
0.211
|
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr11_-_59436366
|
0.211
|
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast)
|
|
chr3_+_122399665
|
0.211
|
NM_017554
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14
|
|
chr1_+_24104906
|
0.210
|
|
PITHD1
|
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1
|
|
chr1_+_3541550
|
0.210
|
NM_182752
|
TPRG1L
|
tumor protein p63 regulated 1-like
|
|
chr9_-_21994379
|
0.210
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr19_-_18654376
|
0.210
|
NM_012181
|
FKBP8
|
FK506 binding protein 8, 38kDa
|
|
chr17_-_7590731
|
0.209
|
NM_000546
NM_001126112
NM_001126113
NM_001126114
|
TP53
|
tumor protein p53
|
|
chr19_-_54984341
|
0.208
|
NM_145057
|
CDC42EP5
|
CDC42 effector protein (Rho GTPase binding) 5
|
|
chr1_-_17307107
|
0.207
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chrX_+_103031753
|
0.207
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr14_+_24867926
|
0.206
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr2_-_72371188
|
0.205
|
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr2_+_30369800
|
0.205
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr3_-_52008645
|
0.204
|
NM_001146314
NM_032750
|
ABHD14B
|
abhydrolase domain containing 14B
|
|
chr2_+_24346339
|
0.204
|
NM_001145710
|
LOC375190
|
UPF0638 protein B
|
|
chrX_+_152760452
|
0.203
|
|
BGN
|
biglycan
|
|
chr1_-_38471170
|
0.202
|
NM_001243878
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chrX_+_152760346
|
0.201
|
NM_001711
|
BGN
|
biglycan
|
|
chr5_-_81046858
|
0.201
|
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chrX_+_152990322
|
0.200
|
NM_000033
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr2_+_30369868
|
0.198
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr19_+_44080951
|
0.198
|
NM_001193621
|
LOC390940
|
uncharacterized protein ENSP00000244321
|
|
chr16_+_20911987
|
0.197
|
NM_001128301
NM_001128302
|
LYRM1
|
LYR motif containing 1
|
|
chr19_+_39279849
|
0.197
|
NM_001042507
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B
|
|
chr19_-_55658310
|
0.197
|
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr7_-_28220146
|
0.196
|
|
JAZF1
|
JAZF zinc finger 1
|
|
chr10_-_103535656
|
0.196
|
NM_006119
NM_033163
NM_033164
NM_033165
|
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chr1_+_3541587
|
0.196
|
|
TPRG1L
|
tumor protein p63 regulated 1-like
|
|
chrX_-_154114431
|
0.196
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr21_+_46825001
|
0.195
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr1_-_6321034
|
0.194
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr1_-_144520987
|
0.194
|
|
LOC728855
|
uncharacterized LOC728855
|
|
chr2_-_68547018
|
0.193
|
NM_001111101
NM_015463
|
CNRIP1
|
cannabinoid receptor interacting protein 1
|
|
chr17_+_55333843
|
0.193
|
NM_138962
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr5_+_139028483
|
0.192
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr12_+_79258794
|
0.192
|
|
SYT1
|
synaptotagmin I
|
|
chr7_-_20826503
|
0.191
|
NM_182700
NM_198956
|
SP8
|
Sp8 transcription factor
|
|
chr2_-_145090038
|
0.191
|
NM_001164629
|
GTDC1
|
glycosyltransferase-like domain containing 1
|