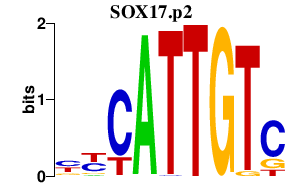
|
chr8_+_81398416
|
0.547
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr2_+_70142236
|
0.394
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr12_-_24103953
|
0.379
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr2_+_70142303
|
0.377
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr2_+_75061234
|
0.370
|
|
HK2
|
hexokinase 2
|
|
chr5_-_64777703
|
0.325
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr3_+_31574119
|
0.324
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr7_-_131241321
|
0.314
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr7_+_69064316
|
0.271
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr17_+_26646120
|
0.266
|
NM_014573
|
TMEM97
|
transmembrane protein 97
|
|
chr8_+_26435590
|
0.241
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr5_+_139781395
|
0.239
|
NM_020690
NM_001197030
NM_017747
NM_017978
NM_024668
|
ANKHD1-EIF4EBP3
ANKHD1
|
ANKHD1-EIF4EBP3 readthrough
ankyrin repeat and KH domain containing 1
|
|
chr5_+_139781450
|
0.234
|
|
ANKHD1-EIF4EBP3
ANKHD1
|
ANKHD1-EIF4EBP3 readthrough
ankyrin repeat and KH domain containing 1
|
|
chr7_+_121513158
|
0.232
|
NM_001206838
NM_001206839
NM_002851
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr16_+_838634
|
0.223
|
|
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr5_+_139781472
|
0.222
|
|
ANKHD1-EIF4EBP3
ANKHD1
|
ANKHD1-EIF4EBP3 readthrough
ankyrin repeat and KH domain containing 1
|
|
chr16_+_838610
|
0.222
|
NM_022092
|
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr16_-_30934437
|
0.221
|
|
FBXL19-AS1
|
FBXL19 antisense RNA 1 (non-protein coding)
|
|
chr20_-_17662704
|
0.217
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr20_+_18794369
|
0.217
|
NM_178483
|
C20orf79
|
chromosome 20 open reading frame 79
|
|
chr19_+_13906207
|
0.214
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr5_+_108084638
|
0.213
|
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr16_+_838645
|
0.213
|
|
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr5_+_139781424
|
0.210
|
|
ANKHD1
|
ankyrin repeat and KH domain containing 1
|
|
chr7_+_116660291
|
0.209
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr14_+_103800492
|
0.208
|
NM_001969
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr19_+_45394459
|
0.206
|
NM_001128916
NM_001128917
NM_006114
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr5_+_102465256
|
0.203
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr20_-_17662828
|
0.202
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr13_-_31040035
|
0.198
|
NM_002128
|
HMGB1
|
high mobility group box 1
|
|
chr4_-_80994309
|
0.187
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr15_+_40886217
|
0.186
|
|
CASC5
|
cancer susceptibility candidate 5
|
|
chr17_-_79196702
|
0.185
|
NM_001009811
NM_014984
|
AZI1
|
5-azacytidine induced 1
|
|
chrX_+_78003205
|
0.184
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr7_+_116660263
|
0.180
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr3_-_20227618
|
0.179
|
NM_001012409
NM_001012410
NM_001012411
NM_001012412
NM_001012413
NM_138484
NM_001199251
NM_001199252
NM_001199253
NM_001199254
NM_001199255
NM_001199256
NM_001199257
|
SGOL1
|
shugoshin-like 1 (S. pombe)
|
|
chr3_+_51422716
|
0.179
|
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr11_-_67141647
|
0.178
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr1_-_8931634
|
0.176
|
NM_001201483
|
ENO1
|
enolase 1, (alpha)
|
|
chr2_+_172950207
|
0.175
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr20_-_56285576
|
0.174
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr2_-_97405775
|
0.172
|
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr12_+_108909049
|
0.168
|
NM_007076
|
FICD
|
FIC domain containing
|
|
chr3_-_48601200
|
0.168
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr4_-_80993713
|
0.167
|
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr5_+_148206155
|
0.167
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr2_+_169757749
|
0.165
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr2_-_97405799
|
0.164
|
NM_001142292
NM_030805
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr16_+_56817367
|
0.162
|
NM_001242796
|
NUP93
|
nucleoporin 93kDa
|
|
chr19_+_3880675
|
0.160
|
|
ATCAY
|
ataxia, cerebellar, Cayman type
|
|
chr6_-_108279307
|
0.160
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr3_+_51422691
|
0.160
|
NM_006010
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr20_+_37554954
|
0.159
|
NM_030919
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr6_+_12290528
|
0.158
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chrX_+_151999605
|
0.156
|
|
NSDHL
|
NAD(P) dependent steroid dehydrogenase-like
|
|
chr13_-_31039966
|
0.153
|
|
HMGB1
|
high mobility group box 1
|
|
chrX_+_48367346
|
0.150
|
NM_022825
NM_203473
|
PORCN
|
porcupine homolog (Drosophila)
|
|
chr1_-_16161279
|
0.150
|
|
|
|
|
chr5_+_140474198
|
0.149
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr1_+_45805877
|
0.148
|
|
TOE1
|
target of EGR1, member 1 (nuclear)
|
|
chr7_-_111846384
|
0.146
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr5_+_68462948
|
0.146
|
|
CCNB1
|
cyclin B1
|
|
chr17_-_61777478
|
0.146
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr11_-_4599049
|
0.146
|
NM_144663
|
C11orf40
|
chromosome 11 open reading frame 40
|
|
chr17_-_8113862
|
0.145
|
NM_004217
|
AURKB
|
aurora kinase B
|
|
chr14_+_103800534
|
0.143
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr3_+_67048726
|
0.142
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr11_+_560868
|
0.142
|
NM_001143993
|
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7
|
|
chr2_-_61765394
|
0.141
|
NM_003400
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr9_-_128003634
|
0.141
|
NM_005347
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr22_+_38142245
|
0.140
|
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr20_+_42086533
|
0.139
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr19_+_45394522
|
0.139
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr1_+_179262936
|
0.139
|
|
SOAT1
|
sterol O-acyltransferase 1
|
|
chr3_-_149510609
|
0.139
|
NM_001144960
|
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1
|
|
chr1_+_27719151
|
0.138
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr4_+_75310852
|
0.137
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr17_-_49124128
|
0.137
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr7_+_89843133
|
0.135
|
NM_001040666
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr11_-_133715368
|
0.134
|
NM_174927
|
SPATA19
|
spermatogenesis associated 19
|
|
chr1_+_43824598
|
0.133
|
NM_001255
|
CDC20
|
cell division cycle 20 homolog (S. cerevisiae)
|
|
chr13_-_31736031
|
0.132
|
NM_006644
|
HSPH1
|
heat shock 105kDa/110kDa protein 1
|
|
chr14_-_106926393
|
0.132
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr1_+_45805783
|
0.132
|
|
TOE1
|
target of EGR1, member 1 (nuclear)
|
|
chr20_+_37555055
|
0.131
|
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr3_+_157154579
|
0.129
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr7_+_150690838
|
0.129
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chrX_+_151999565
|
0.128
|
|
NSDHL
|
NAD(P) dependent steroid dehydrogenase-like
|
|
chrX_+_151999510
|
0.127
|
NM_001129765
NM_015922
|
NSDHL
|
NAD(P) dependent steroid dehydrogenase-like
|
|
chr5_+_68462985
|
0.127
|
|
CCNB1
|
cyclin B1
|
|
chr2_+_197504277
|
0.127
|
NM_001080539
|
CCDC150
|
coiled-coil domain containing 150
|
|
chr1_-_197115566
|
0.126
|
NM_001206846
NM_018136
|
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila)
|
|
chr14_+_85996512
|
0.126
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chrX_-_47931000
|
0.125
|
NM_001190255
|
ZNF630
|
zinc finger protein 630
|
|
chr1_+_45805669
|
0.124
|
|
TOE1
|
target of EGR1, member 1 (nuclear)
|
|
chr14_+_85996454
|
0.124
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr9_+_134000939
|
0.124
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr6_+_25754926
|
0.123
|
NM_005495
|
SLC17A4
|
solute carrier family 17 (sodium phosphate), member 4
|
|
chr6_-_139613114
|
0.123
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr2_-_61765345
|
0.123
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr11_+_33563772
|
0.121
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr6_-_108279212
|
0.120
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr12_-_122985471
|
0.119
|
NM_017612
|
ZCCHC8
|
zinc finger, CCHC domain containing 8
|
|
chr19_+_50031232
|
0.119
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr3_-_137851123
|
0.118
|
NM_016161
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase
|
|
chr2_-_220094354
|
0.118
|
NM_001077198
NM_024085
|
ATG9A
|
ATG9 autophagy related 9 homolog A (S. cerevisiae)
|
|
chr5_+_176875325
|
0.118
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr2_-_61765367
|
0.118
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr15_+_90118740
|
0.117
|
NM_152259
|
C15orf42
|
chromosome 15 open reading frame 42
|
|
chr4_+_2061238
|
0.117
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr16_-_2264776
|
0.116
|
NM_001042371
|
PGP
|
phosphoglycolate phosphatase
|
|
chr3_-_9439122
|
0.115
|
|
LOC440944
|
uncharacterized LOC440944
|
|
chr1_+_179262960
|
0.115
|
|
SOAT1
|
sterol O-acyltransferase 1
|
|
chr6_+_80714389
|
0.115
|
|
TTK
|
TTK protein kinase
|
|
chr9_+_130159383
|
0.115
|
NM_014580
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8
|
|
chr14_+_23938902
|
0.114
|
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr17_-_38210611
|
0.114
|
|
MED24
|
mediator complex subunit 24
|
|
chr20_-_13971256
|
0.113
|
NM_025229
|
SEL1L2
|
sel-1 suppressor of lin-12-like 2 (C. elegans)
|
|
chr6_-_53213586
|
0.113
|
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chr11_+_128563772
|
0.112
|
NM_001167681
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr12_-_6960406
|
0.111
|
NM_031299
|
CDCA3
|
cell division cycle associated 3
|
|
chr1_+_46713385
|
0.111
|
|
RAD54L
|
RAD54-like (S. cerevisiae)
|
|
chr3_+_45927995
|
0.111
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr1_+_201709051
|
0.110
|
|
NAV1
|
neuron navigator 1
|
|
chr17_-_48262900
|
0.110
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr2_+_73612821
|
0.110
|
NM_015120
|
ALMS1
|
Alstrom syndrome 1
|
|
chr17_+_57232859
|
0.109
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr1_+_45805341
|
0.109
|
NM_025077
|
TOE1
|
target of EGR1, member 1 (nuclear)
|
|
chr5_+_145826866
|
0.109
|
NM_001040006
NM_006706
|
TCERG1
|
transcription elongation regulator 1
|
|
chr9_-_128003348
|
0.108
|
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr1_+_46713359
|
0.108
|
NM_001142548
NM_003579
|
RAD54L
|
RAD54-like (S. cerevisiae)
|
|
chr11_-_28129687
|
0.107
|
|
KIF18A
|
kinesin family member 18A
|
|
chr11_-_28129646
|
0.106
|
|
KIF18A
|
kinesin family member 18A
|
|
chr5_-_16936371
|
0.105
|
NM_012334
|
MYO10
|
myosin X
|
|
chr2_-_174767282
|
0.105
|
|
|
|
|
chr9_-_74383301
|
0.104
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr1_-_33815498
|
0.104
|
NM_004427
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr4_+_55523906
|
0.104
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr15_+_39873276
|
0.104
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr11_-_28129701
|
0.104
|
NM_031217
|
KIF18A
|
kinesin family member 18A
|
|
chr2_+_153191830
|
0.104
|
|
FMNL2
|
formin-like 2
|
|
chr7_-_92463211
|
0.103
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr14_+_103800678
|
0.103
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr9_-_128003615
|
0.103
|
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr8_-_82024277
|
0.102
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr14_+_103800548
|
0.102
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr9_-_99180661
|
0.102
|
NM_153695
|
ZNF367
|
zinc finger protein 367
|
|
chr2_+_10262694
|
0.102
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr9_+_140135738
|
0.101
|
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chr2_+_10262853
|
0.101
|
NM_001034
|
RRM2
|
ribonucleotide reductase M2
|
|
chr5_+_112312427
|
0.101
|
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr1_-_53018721
|
0.101
|
NM_001009881
NM_015269
|
ZCCHC11
|
zinc finger, CCHC domain containing 11
|
|
chr3_+_5229430
|
0.101
|
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1
|
|
chr15_+_40886442
|
0.100
|
NM_144508
NM_170589
|
CASC5
|
cancer susceptibility candidate 5
|
|
chr14_-_35344450
|
0.100
|
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr14_-_107218786
|
0.100
|
|
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr21_-_16437125
|
0.100
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr2_+_54683276
|
0.100
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr6_-_108395939
|
0.100
|
NM_014028
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr4_-_80994183
|
0.099
|
|
|
|
|
chr14_-_60097223
|
0.099
|
|
RTN1
|
reticulon 1
|
|
chr6_-_62995852
|
0.098
|
NM_152688
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2
|
|
chr5_-_137548982
|
0.098
|
|
CDC23
|
cell division cycle 23 homolog (S. cerevisiae)
|
|
chr3_+_1134619
|
0.098
|
NM_014461
|
CNTN6
|
contactin 6
|
|
chr1_+_45792544
|
0.098
|
NM_032756
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase-like
|
|
chr12_-_52585686
|
0.097
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr19_+_4247049
|
0.097
|
NM_018074
|
CCDC94
|
coiled-coil domain containing 94
|
|
chr5_-_24645084
|
0.097
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr7_-_25164902
|
0.097
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr3_-_192635933
|
0.097
|
NM_178496
|
MB21D2
|
Mab-21 domain containing 2
|
|
chrX_+_70443055
|
0.096
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr18_+_31158540
|
0.096
|
NM_030632
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr14_-_35344080
|
0.096
|
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr20_-_590909
|
0.095
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr9_-_35828728
|
0.094
|
NM_001012446
|
C9orf128
|
chromosome 9 open reading frame 128
|
|
chr9_+_140135698
|
0.094
|
NM_006088
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chr17_+_38444130
|
0.094
|
|
CDC6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr11_+_13690151
|
0.094
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr12_-_75905382
|
0.093
|
|
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast)
|
|
chr22_+_26825220
|
0.092
|
NM_020437
|
ASPHD2
|
aspartate beta-hydroxylase domain containing 2
|
|
chr3_+_5229357
|
0.092
|
NM_014674
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1
|
|
chr17_-_61777467
|
0.092
|
|
LIMD2
|
LIM domain containing 2
|
|
chr15_+_77223942
|
0.092
|
NM_002902
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain
|
|
chr15_+_89164522
|
0.092
|
NM_022767
|
AEN
|
apoptosis enhancing nuclease
|
|
chr20_+_207898
|
0.092
|
NM_080831
|
DEFB129
|
defensin, beta 129
|
|
chr5_-_77805930
|
0.091
|
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr11_-_67141008
|
0.091
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr8_-_117023
|
0.091
|
NM_001005504
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21
|
|
chr1_-_43424324
|
0.091
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr17_+_38444134
|
0.090
|
NM_001254
|
CDC6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr9_+_140135742
|
0.090
|
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chr12_-_45270071
|
0.090
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr11_-_46867788
|
0.090
|
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr1_+_46713433
|
0.090
|
|
RAD54L
|
RAD54-like (S. cerevisiae)
|
|
chr16_-_81253974
|
0.090
|
NM_001076780
NM_052892
|
PKD1L2
|
polycystic kidney disease 1-like 2
|
|
chr6_-_108279320
|
0.090
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr13_-_31735718
|
0.089
|
|
HSPH1
|
heat shock 105kDa/110kDa protein 1
|
|
chr15_+_40650411
|
0.089
|
NM_033510
|
DISP2
|
dispatched homolog 2 (Drosophila)
|
|
chr4_+_154387450
|
0.089
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr16_+_56815703
|
0.089
|
NM_001242795
|
NUP93
|
nucleoporin 93kDa
|