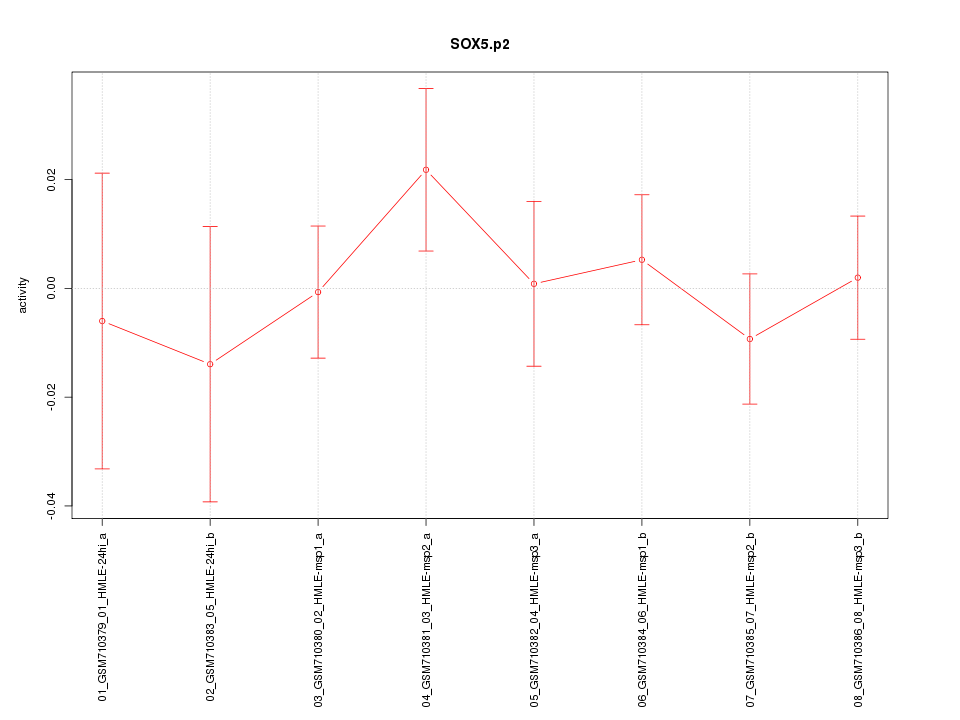
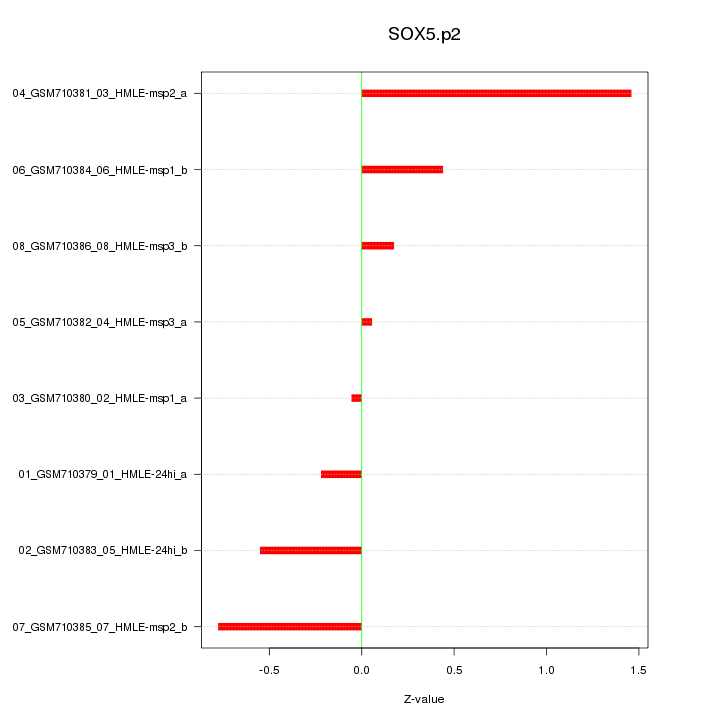
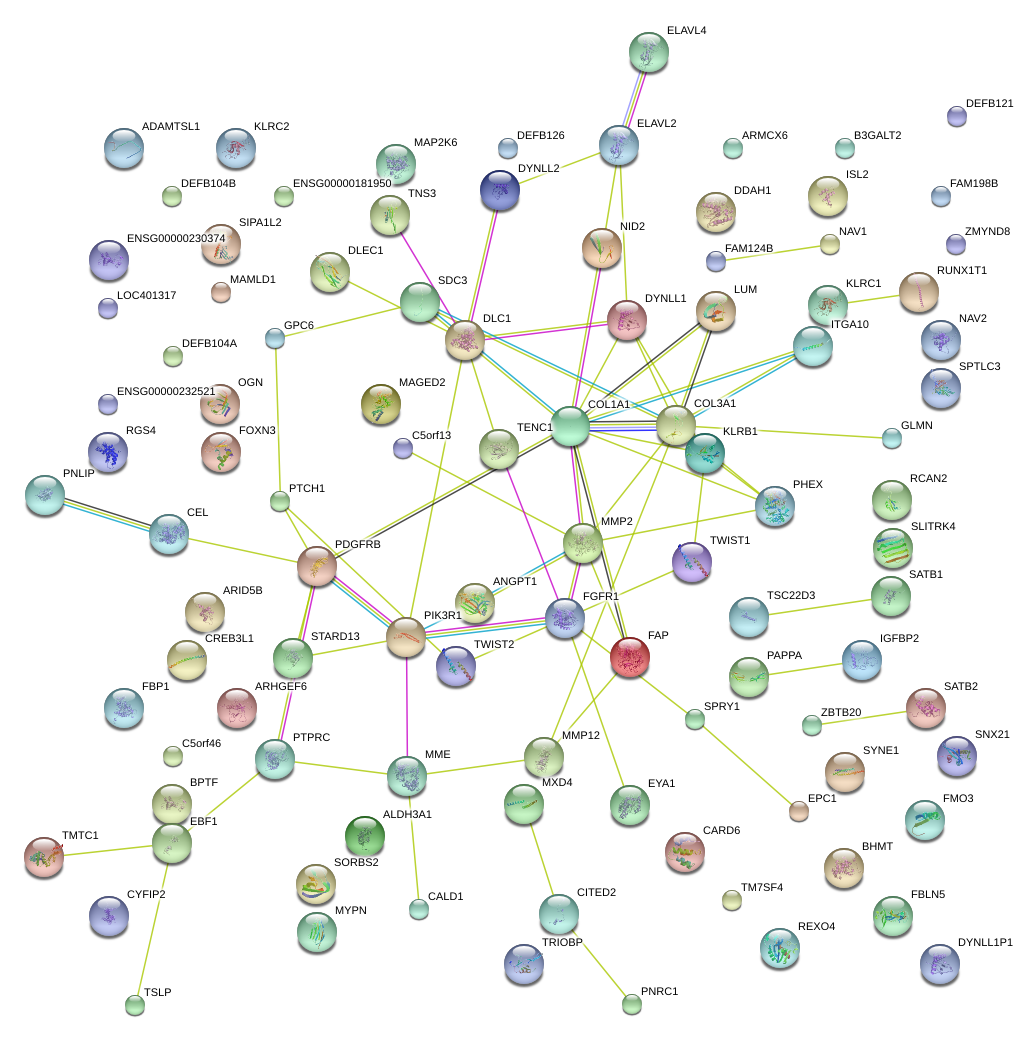
|
chr14_-_92413738
|
1.770
|
|
FBLN5
|
fibulin 5
|
|
chr14_-_92414004
|
1.392
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr1_+_163038564
|
1.140
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr2_+_189839132
|
0.799
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr8_-_108510094
|
0.760
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr3_-_114790221
|
0.674
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr6_-_46293404
|
0.600
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr5_-_111312522
|
0.541
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_163038934
|
0.527
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr5_-_147286064
|
0.523
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr9_-_95166827
|
0.515
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr5_+_110407389
|
0.466
|
NM_033035
|
TSLP
|
thymic stromal lymphopoietin
|
|
chr8_-_93075190
|
0.431
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_+_67588395
|
0.407
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr5_-_111092918
|
0.395
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_158526698
|
0.383
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr5_-_111093251
|
0.351
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr4_+_124317884
|
0.349
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr12_-_29937691
|
0.341
|
NM_175861
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr5_-_111093030
|
0.334
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_-_163099877
|
0.331
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr3_-_151987332
|
0.327
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr1_-_232651227
|
0.325
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chrX_+_149613719
|
0.313
|
NM_001177466
NM_005491
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr5_-_111092865
|
0.313
|
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_+_156820161
|
0.312
|
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr4_-_159093523
|
0.311
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr12_-_91505215
|
0.310
|
|
LUM
|
lumican
|
|
chr17_-_19648924
|
0.307
|
NM_001135167
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr3_+_154797644
|
0.305
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr12_-_91505541
|
0.301
|
NM_002345
|
LUM
|
lumican
|
|
chr5_-_111091947
|
0.301
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr8_-_93115453
|
0.299
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr20_-_29993955
|
0.296
|
NM_001011878
|
DEFB121
|
defensin, beta 121
|
|
chr6_-_139695784
|
0.293
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr12_+_53443834
|
0.292
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr3_+_154797435
|
0.291
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr8_-_38325241
|
0.291
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr17_+_65822206
|
0.280
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr8_-_38326115
|
0.280
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr8_-_38325524
|
0.278
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr8_+_7693992
|
0.270
|
NM_080389
NM_001040702
|
DEFB104A
DEFB104B
|
defensin, beta 104A
defensin, beta 104B
|
|
chr4_-_2263808
|
0.267
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr8_-_13134028
|
0.264
|
|
DLC1
|
deleted in liver cancer 1
|
|
chr6_-_139695739
|
0.263
|
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr1_-_85930610
|
0.262
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr12_-_10605928
|
0.260
|
NM_002259
NM_007328
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr4_-_159093717
|
0.256
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_-_2263706
|
0.255
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr6_-_139695349
|
0.251
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chrX_-_142723921
|
0.250
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chrX_-_135749436
|
0.248
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr9_-_97402530
|
0.246
|
NM_001127628
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr8_-_93029864
|
0.244
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr10_+_118305427
|
0.230
|
NM_000936
|
PNLIP
|
pancreatic lipase
|
|
chr20_+_123230
|
0.229
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr13_-_33760215
|
0.228
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr7_-_47622127
|
0.226
|
|
TNS3
|
tensin 3
|
|
chr1_+_171060017
|
0.225
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr8_-_72274466
|
0.225
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr9_-_98268866
|
0.222
|
|
PTCH1
|
patched 1
|
|
chr22_+_38092994
|
0.220
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr9_+_118916069
|
0.220
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr20_+_44462395
|
0.218
|
NM_001042632
NM_001042633
NM_033421
NM_152897
|
SNX21
|
sorting nexin family member 21
|
|
chr14_-_89883306
|
0.217
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr17_-_48264039
|
0.214
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr3_-_18480203
|
0.213
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr1_-_86043932
|
0.213
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr17_+_65822242
|
0.212
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr1_-_31345361
|
0.212
|
|
SDC3
|
syndecan 3
|
|
chr4_-_186732208
|
0.211
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr15_+_76629101
|
0.209
|
NM_145805
|
ISL2
|
ISL LIM homeobox 2
|
|
chr8_+_105352050
|
0.207
|
NM_030788
|
TM7SF4
|
transmembrane 7 superfamily member 4
|
|
chr5_-_149535138
|
0.206
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr10_-_32667543
|
0.206
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr9_+_18474078
|
0.205
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr20_-_45985411
|
0.204
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr6_-_152957985
|
0.202
|
NM_033071
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr2_-_200325200
|
0.201
|
|
SATB2
|
SATB homeobox 2
|
|
chrX_+_22050920
|
0.199
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chrX_+_54834107
|
0.198
|
NM_177433
|
MAGED2
|
melanoma antigen family D, 2
|
|
chr5_+_78407603
|
0.198
|
NM_001713
|
BHMT
|
betaine--homocysteine S-methyltransferase
|
|
chr13_+_93879011
|
0.196
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr20_+_12989616
|
0.196
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr2_-_225266710
|
0.195
|
NM_001122779
NM_024785
|
FAM124B
|
family with sequence similarity 124B
|
|
chr3_-_114343052
|
0.195
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr10_+_69869249
|
0.192
|
NM_032578
|
MYPN
|
myopalladin
|
|
chrX_-_106959597
|
0.188
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr4_-_186732047
|
0.181
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_+_19372270
|
0.179
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr7_+_134576187
|
0.179
|
|
CALD1
|
caldesmon 1
|
|
chr1_+_198608244
|
0.178
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chrX_-_100872926
|
0.176
|
NM_001009584
NM_001184768
NM_019007
|
ARMCX6
|
armadillo repeat containing, X-linked 6
|
|
chr14_-_52535711
|
0.175
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr10_+_63661012
|
0.174
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr11_+_46299554
|
0.173
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr17_+_67498391
|
0.172
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr7_+_28448970
|
0.172
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr5_+_40841296
|
0.170
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr11_+_46299585
|
0.170
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr2_+_217498082
|
0.169
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr1_+_201709051
|
0.169
|
|
NAV1
|
neuron navigator 1
|
|
chr1_+_145524989
|
0.169
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr1_+_50571963
|
0.168
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr7_-_19157262
|
0.167
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr6_+_89790464
|
0.166
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr8_-_72273965
|
0.165
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr6_+_89790412
|
0.165
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr16_+_55515468
|
0.165
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr7_-_47621635
|
0.163
|
|
TNS3
|
tensin 3
|
|
chr1_-_193155723
|
0.161
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr14_-_52535765
|
0.160
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr11_-_102401430
|
0.159
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr16_+_86612114
|
0.159
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr6_+_119215240
|
0.156
|
NM_014034
|
ASF1A
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae)
|
|
chr3_-_165555167
|
0.156
|
|
BCHE
|
butyrylcholinesterase
|
|
chr1_-_156217717
|
0.155
|
NM_198406
NM_024897
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr21_-_28339405
|
0.155
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr18_+_72922725
|
0.154
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr3_+_12330336
|
0.154
|
NM_138711
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr6_+_155538096
|
0.154
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr13_-_74708361
|
0.153
|
|
KLF12
|
Kruppel-like factor 12
|
|
chr1_+_221052735
|
0.152
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr7_-_27142301
|
0.151
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr3_-_122283423
|
0.151
|
NM_001146103
NM_031458
|
PARP9
|
poly (ADP-ribose) polymerase family, member 9
|
|
chr11_+_65192284
|
0.150
|
|
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr5_+_156696351
|
0.149
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr1_-_108231122
|
0.149
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr10_+_31610063
|
0.147
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr6_+_132891460
|
0.147
|
NM_175067
|
TAAR6
|
trace amine associated receptor 6
|
|
chr4_-_175443601
|
0.146
|
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD)
|
|
chr5_-_168271852
|
0.146
|
|
|
|
|
chr7_+_104681378
|
0.146
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr3_+_12392993
|
0.146
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr18_-_52625278
|
0.146
|
NM_001143829
|
CCDC68
|
coiled-coil domain containing 68
|
|
chrX_-_154250769
|
0.145
|
NM_000132
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr7_-_47621741
|
0.145
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr1_+_174844655
|
0.144
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr10_+_99079017
|
0.143
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr4_-_104020964
|
0.143
|
NM_020139
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2
|
|
chr1_+_164528914
|
0.143
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr2_+_109237679
|
0.140
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr5_-_171710794
|
0.140
|
NM_152277
|
UBTD2
|
ubiquitin domain containing 2
|
|
chr14_-_73493744
|
0.139
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr17_+_58677603
|
0.138
|
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr14_-_52535737
|
0.138
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr4_+_113970784
|
0.137
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr8_-_57123827
|
0.137
|
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr22_-_19418998
|
0.136
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr19_+_36132646
|
0.136
|
NM_014209
|
ETV2
|
ets variant 2
|
|
chr10_+_35416089
|
0.135
|
|
CREM
|
cAMP responsive element modulator
|
|
chr1_+_145663156
|
0.135
|
|
RNF115
|
ring finger protein 115
|
|
chr17_+_3100809
|
0.134
|
NM_012352
|
OR1A2
|
olfactory receptor, family 1, subfamily A, member 2
|
|
chrX_+_135251795
|
0.133
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_+_132129195
|
0.133
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr3_+_69985750
|
0.133
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr14_+_53019865
|
0.132
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr5_+_145718586
|
0.132
|
NM_002700
|
POU4F3
|
POU class 4 homeobox 3
|
|
chr1_+_51434447
|
0.132
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr11_+_22696318
|
0.132
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chrX_-_142722869
|
0.132
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr12_-_10607214
|
0.131
|
NM_213657
NM_213658
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr5_+_137203559
|
0.131
|
|
MYOT
|
myotilin
|
|
chr2_-_175869910
|
0.130
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr4_+_86699858
|
0.129
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr8_-_38325825
|
0.129
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr1_+_114522029
|
0.129
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr2_-_188312872
|
0.128
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr16_-_67977956
|
0.128
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr12_-_31743822
|
0.128
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr5_+_15500288
|
0.128
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr4_+_8951476
|
0.128
|
NM_001040071
|
LOC650293
|
seven transmembrane helix receptor
|
|
chr7_-_111846384
|
0.127
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr5_+_137203544
|
0.127
|
NM_001135940
NM_006790
|
MYOT
|
myotilin
|
|
chr1_-_182360400
|
0.126
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr16_+_69726751
|
0.125
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr8_-_57123858
|
0.125
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr3_-_187871816
|
0.125
|
|
LPP-AS2
|
LPP antisense RNA 2 (non-protein coding)
|
|
chr5_-_154230157
|
0.124
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr11_-_4599049
|
0.124
|
NM_144663
|
C11orf40
|
chromosome 11 open reading frame 40
|
|
chr4_+_86851425
|
0.124
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr4_+_55095433
|
0.124
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr13_-_33780142
|
0.123
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr22_-_19419215
|
0.123
|
NM_003325
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr4_+_160188997
|
0.123
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr7_+_129932973
|
0.123
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr22_-_19419177
|
0.122
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr5_+_140782367
|
0.121
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr16_+_69680959
|
0.119
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr3_-_66551341
|
0.119
|
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr2_-_128421965
|
0.119
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr5_-_95158465
|
0.117
|
NM_001118890
NM_001243658
NM_001243659
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr3_+_46616044
|
0.115
|
NM_001174136
|
TDGF1
|
teratocarcinoma-derived growth factor 1
|
|
chr5_-_38595506
|
0.115
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chrX_+_107683015
|
0.113
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr3_-_99594915
|
0.113
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr3_-_48754589
|
0.112
|
NM_001005909
NM_001005910
NM_001005911
NM_001146178
NM_001146179
NM_001190316
NM_001190317
NM_016291
|
IP6K2
|
inositol hexakisphosphate kinase 2
|
|
chr1_+_33722158
|
0.112
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr1_-_163113129
|
0.111
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr1_+_170633312
|
0.110
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|