|
chr5_+_71502700
|
0.303
|
|
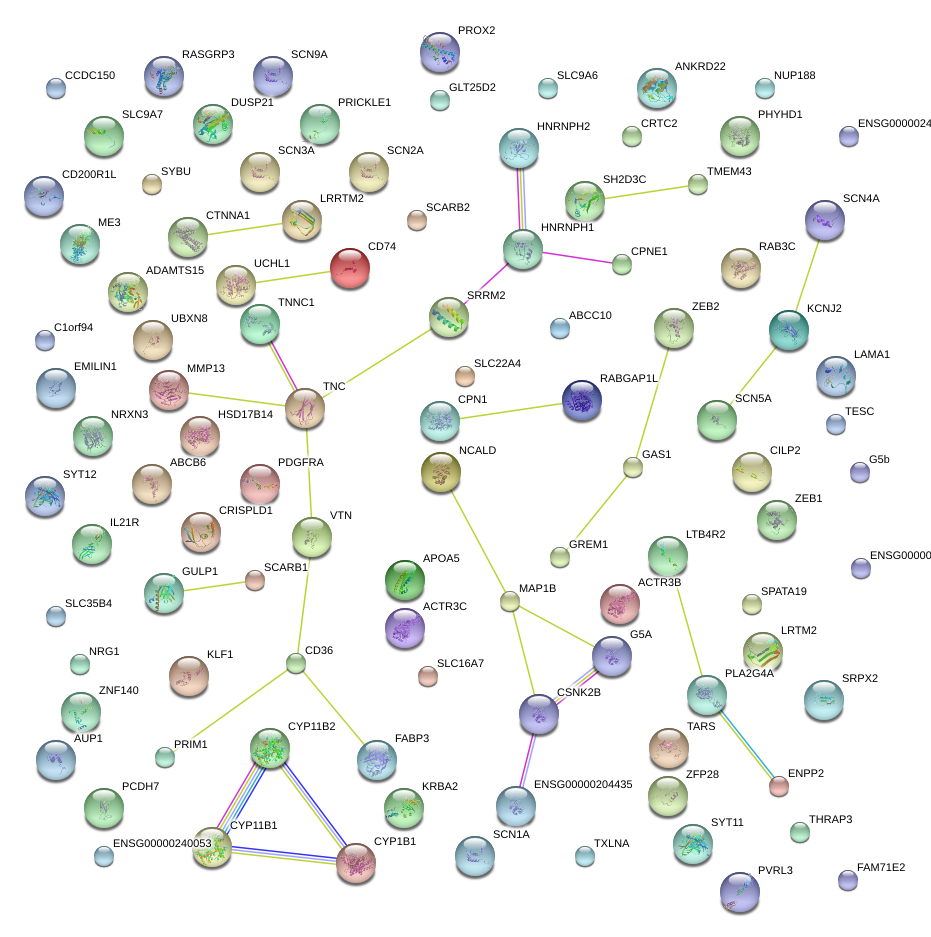
MAP1B
|
microtubule-associated protein 1B
|
|
chr11_-_133715368
|
0.251
|
NM_174927
|
SPATA19
|
spermatogenesis associated 19
|
|
chr3_-_112564789
|
0.201
|
NM_001008784
NM_001199215
|
CD200R1L
|
CD200 receptor 1-like
|
|
chr9_-_89562103
|
0.181
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr4_+_41258895
|
0.170
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr4_+_41258909
|
0.166
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr1_-_31845818
|
0.163
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr4_+_41258941
|
0.158
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr14_+_24779356
|
0.151
|
NM_001164692
NM_019839
|
LTB4R2
|
leukotriene B4 receptor 2
|
|
chr2_-_38303265
|
0.138
|
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr12_-_42877785
|
0.137
|
NM_001144882
NM_001144883
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr1_+_186798031
|
0.135
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr8_+_30601666
|
0.135
|
NM_005671
|
UBXN8
|
UBX domain protein 8
|
|
chr2_+_33701320
|
0.131
|
NM_001139488
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr12_-_42877743
|
0.130
|
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr5_+_57878901
|
0.129
|
NM_138453
|
RAB3C
|
RAB3C, member RAS oncogene family
|
|
chr5_-_138210992
|
0.124
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr4_+_55095263
|
0.123
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr15_+_33010174
|
0.120
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr3_+_14166552
|
0.118
|
|
TMEM43
|
transmembrane protein 43
|
|
chr11_-_102826433
|
0.116
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr3_+_14166543
|
0.115
|
|
TMEM43
|
transmembrane protein 43
|
|
chr3_+_14166582
|
0.113
|
|
TMEM43
|
transmembrane protein 43
|
|
chr5_-_138211054
|
0.113
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr1_-_153931077
|
0.110
|
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr3_+_14166439
|
0.110
|
NM_024334
|
TMEM43
|
transmembrane protein 43
|
|
chr21_+_26934440
|
0.109
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr11_+_130318457
|
0.108
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr19_+_57050299
|
0.103
|
NM_020828
|
ZFP28
|
zinc finger protein 28 homolog (mouse)
|
|
chr3_+_14166632
|
0.101
|
|
TMEM43
|
transmembrane protein 43
|
|
chr8_+_32405727
|
0.100
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr16_+_2802634
|
0.099
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr2_+_27301434
|
0.099
|
NM_007046
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr11_-_86383653
|
0.099
|
NM_001161586
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr1_+_36690038
|
0.098
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr9_+_127420690
|
0.094
|
|
MIR181A2HG
|
MIR181A2 host gene (non-protein coding)
|
|
chr16_+_2802656
|
0.091
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr16_+_2802605
|
0.089
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr16_+_2802683
|
0.087
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr8_-_120651019
|
0.087
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr4_+_30722445
|
0.087
|
|
PCDH7
|
protocadherin 7
|
|
chr1_+_155853546
|
0.086
|
|
SYT11
|
synaptotagmin XI
|
|
chr16_+_2802673
|
0.081
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr10_-_101841593
|
0.080
|
NM_001308
|
CPN1
|
carboxypeptidase N, polypeptide 1
|
|
chr8_-_110661007
|
0.080
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr2_+_27301693
|
0.079
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr2_-_220083699
|
0.078
|
NM_005689
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr9_-_117880411
|
0.078
|
NM_002160
|
TNC
|
tenascin C
|
|
chr6_+_43399488
|
0.078
|
NM_033450
|
ABCC10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10
|
|
chr10_-_90611675
|
0.076
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr17_+_68165660
|
0.076
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr7_-_134001502
|
0.076
|
|
SLC35B4
|
solute carrier family 35, member B4
|
|
chr17_-_1619473
|
0.076
|
|
MIR22HG
|
MIR22 host gene (non-protein coding)
|
|
chr2_-_38303307
|
0.075
|
NM_000104
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr9_-_130517552
|
0.075
|
NM_001142534
NM_001142533
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr16_+_2802708
|
0.074
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr7_-_97601654
|
0.074
|
|
|
|
|
chr16_+_27413482
|
0.074
|
NM_181078
NM_181079
|
IL21R
|
interleukin 21 receptor
|
|
chr2_-_74756823
|
0.073
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr12_+_1929432
|
0.073
|
NM_001039029
NM_001163925
NM_001163926
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr2_-_145277568
|
0.073
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr11_-_116663106
|
0.073
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr19_-_12997956
|
0.072
|
NM_006563
|
KLF1
|
Kruppel-like factor 1 (erythroid)
|
|
chr12_+_60083125
|
0.071
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr2_-_74756838
|
0.070
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr7_+_80275634
|
0.070
|
NM_001127443
NM_001127444
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr14_+_78870092
|
0.070
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr12_+_133656984
|
0.069
|
NM_003440
|
ZNF140
|
zinc finger protein 140
|
|
chr2_+_189157551
|
0.068
|
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr3_+_110790460
|
0.068
|
NM_001243286
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chrX_+_44703248
|
0.067
|
NM_022076
|
DUSP21
|
dual specificity phosphatase 21
|
|
chr7_-_134001749
|
0.066
|
NM_032826
|
SLC35B4
|
solute carrier family 35, member B4
|
|
chr1_-_184006828
|
0.066
|
NM_015101
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chrM_+_6138
|
0.066
|
|
|
|
|
chr5_-_149792370
|
0.064
|
NM_001025158
NM_001025159
NM_004355
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr2_-_74756755
|
0.063
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr12_-_46384208
|
0.063
|
|
SCAF11
|
SR-related CTD-associated factor 11
|
|
chr5_+_33440801
|
0.063
|
NM_152295
|
TARS
|
threonyl-tRNA synthetase
|
|
chr6_+_31638727
|
0.063
|
NM_021221
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B
|
|
chr5_+_131630065
|
0.063
|
NM_003059
|
SLC22A4
|
solute carrier family 22 (organic cation/ergothioneine transporter), member 4
|
|
chr7_-_150020661
|
0.062
|
NM_001164458
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast)
|
|
chr12_-_117537090
|
0.062
|
NM_001168325
NM_017899
|
TESC
|
tescalcin
|
|
chr2_-_166060552
|
0.062
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr2_-_74756824
|
0.061
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr2_+_197504277
|
0.061
|
NM_001080539
|
CCDC150
|
coiled-coil domain containing 150
|
|
chr2_-_74756815
|
0.061
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr2_-_167232464
|
0.059
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr1_+_32645322
|
0.059
|
NM_175852
|
TXLNA
|
taxilin alpha
|
|
chrX_+_99899388
|
0.059
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr1_+_34632483
|
0.059
|
NM_032884
|
C1orf94
|
chromosome 1 open reading frame 94
|
|
chr8_+_75896707
|
0.059
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr19_-_49339913
|
0.058
|
NM_016246
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14
|
|
chrX_+_100663205
|
0.058
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr18_-_7117757
|
0.057
|
NM_005559
|
LAMA1
|
laminin, alpha 1
|
|
chr1_+_174844655
|
0.057
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chrX_+_135067573
|
0.057
|
NM_001177651
NM_001042537
NM_006359
|
SLC9A6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr12_-_57146081
|
0.057
|
NM_000946
|
PRIM1
|
primase, DNA, polypeptide 1 (49kDa)
|
|
chr17_-_8274852
|
0.057
|
NM_213597
|
KRBA2
|
KRAB-A domain containing 2
|
|
chr17_-_26697224
|
0.056
|
|
VTN
|
vitronectin
|
|
chr9_+_131683173
|
0.056
|
NM_001100876
NM_174933
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr18_-_56296188
|
0.056
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr2_-_31361591
|
0.055
|
NM_001253826
NM_024572
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr19_-_12845539
|
0.055
|
|
C19orf43
|
chromosome 19 open reading frame 43
|
|
chr1_-_153930911
|
0.055
|
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr19_+_50192440
|
0.055
|
|
C19orf76
|
chromosome 19 open reading frame 76
|
|
chr19_+_50191920
|
0.055
|
NM_001101340
|
C19orf76
|
chromosome 19 open reading frame 76
|
|
chr2_+_172864803
|
0.054
|
NM_199227
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial)
|
|
chr17_-_27053119
|
0.054
|
NM_138463
|
TLCD1
|
TLC domain containing 1
|
|
chr2_-_178937472
|
0.054
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr11_+_65271145
|
0.054
|
|
|
|
|
chr12_-_110434028
|
0.054
|
|
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2
|
|
chrX_+_119495939
|
0.053
|
NM_001142447
NM_012069
|
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide
|
|
chr12_-_11214892
|
0.053
|
NM_176887
|
TAS2R46
|
taste receptor, type 2, member 46
|
|
chr14_-_96000962
|
0.053
|
|
SNHG10
|
small nucleolar RNA host gene 10 (non-protein coding)
|
|
chr17_+_32582295
|
0.053
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr12_-_8380193
|
0.053
|
NM_018088
|
FAM90A1
|
family with sequence similarity 90, member A1
|
|
chr2_+_177016112
|
0.053
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr19_+_55344130
|
0.053
|
NM_012314
|
KIR2DS4
|
killer cell immunoglobulin-like receptor, two domains, short cytoplasmic tail, 4
|
|
chr12_-_93964636
|
0.053
|
|
|
|
|
chr16_-_28550227
|
0.052
|
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr3_-_186079776
|
0.052
|
NM_001080744
NM_001080745
NM_001346
|
DGKG
|
diacylglycerol kinase, gamma 90kDa
|
|
chr6_+_155538096
|
0.052
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr14_-_100841656
|
0.052
|
|
WARS
|
tryptophanyl-tRNA synthetase
|
|
chr14_-_100841793
|
0.052
|
|
WARS
|
tryptophanyl-tRNA synthetase
|
|
chr15_+_71184781
|
0.052
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr9_+_103791030
|
0.052
|
NM_207299
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr7_-_14025826
|
0.051
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr22_-_36019363
|
0.051
|
NM_203377
|
MB
|
myoglobin
|
|
chr11_+_10326618
|
0.051
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr16_+_70207927
|
0.051
|
NM_173619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr1_+_90098643
|
0.051
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr2_+_166150340
|
0.051
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr17_-_53800074
|
0.051
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chrX_+_135067634
|
0.051
|
|
SLC9A6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr1_-_54304107
|
0.051
|
NM_001168551
NM_018087
|
TMEM48
|
transmembrane protein 48
|
|
chr1_-_153931096
|
0.051
|
NM_181715
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr5_-_111092918
|
0.051
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr15_+_62682682
|
0.051
|
|
TLN2
|
talin 2
|
|
chr14_+_38678594
|
0.051
|
|
SSTR1
|
somatostatin receptor 1
|
|
chr9_+_131684561
|
0.050
|
NM_001100877
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr17_-_76356153
|
0.050
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr10_-_12085022
|
0.050
|
NM_080599
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr10_-_121356527
|
0.050
|
NM_001033925
NM_003252
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1
|
|
chr5_-_55290601
|
0.050
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr14_+_32798478
|
0.050
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr9_-_110251308
|
0.050
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr8_+_36641841
|
0.050
|
NM_001031836
|
KCNU1
|
potassium channel, subfamily U, member 1
|
|
chr22_+_20067732
|
0.050
|
NM_001190326
NM_022720
|
DGCR8
|
DiGeorge syndrome critical region gene 8
|
|
chrX_-_34150427
|
0.050
|
NM_203408
|
FAM47A
|
family with sequence similarity 47, member A
|
|
chr12_-_11150473
|
0.049
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr7_+_120629444
|
0.049
|
NM_001105533
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chrX_+_152770083
|
0.049
|
|
BGN
|
biglycan
|
|
chr19_-_49339667
|
0.049
|
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14
|
|
chr16_+_69984625
|
0.049
|
NM_001136214
|
CLEC18A
|
C-type lectin domain family 18, member A
|
|
chr1_-_54303933
|
0.049
|
|
TMEM48
|
transmembrane protein 48
|
|
chr11_+_6866882
|
0.048
|
NM_178168
|
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5
|
|
chr2_+_177016337
|
0.048
|
|
|
|
|
chr4_+_41937105
|
0.048
|
NM_018126
|
TMEM33
|
transmembrane protein 33
|
|
chr8_+_77593522
|
0.048
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr5_+_143550395
|
0.048
|
NM_020768
|
KCTD16
|
potassium channel tetramerisation domain containing 16
|
|
chr17_-_26697310
|
0.048
|
|
VTN
|
vitronectin
|
|
chr19_-_44439410
|
0.048
|
NM_003425
|
ZNF45
|
zinc finger protein 45
|
|
chr8_+_32579350
|
0.048
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr3_+_159706622
|
0.047
|
NM_000882
|
IL12A
|
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35)
|
|
chr12_-_46662779
|
0.047
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chrX_-_110039285
|
0.047
|
NM_001143981
NM_001143982
NM_001143983
|
CHRDL1
|
chordin-like 1
|
|
chr5_+_55062712
|
0.047
|
NM_001166534
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr16_-_28550324
|
0.047
|
NM_001042483
NM_012385
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr14_-_35344852
|
0.047
|
NM_013448
NM_182648
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr4_+_114214102
|
0.046
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chrX_+_69509878
|
0.046
|
NM_012310
|
KIF4A
|
kinesin family member 4A
|
|
chr10_+_92631697
|
0.046
|
NM_001104546
NM_006413
|
RPP30
|
ribonuclease P/MRP 30kDa subunit
|
|
chr6_-_137113624
|
0.046
|
NM_005923
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5
|
|
chr5_-_177423242
|
0.045
|
NM_006261
|
PROP1
|
PROP paired-like homeobox 1
|
|
chr8_+_77593506
|
0.045
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr1_-_53163985
|
0.045
|
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr9_-_110251754
|
0.045
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_-_53164014
|
0.045
|
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr11_-_27681195
|
0.045
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_104893894
|
0.044
|
NM_001136109
NM_001136110
NM_001136112
NM_004347
|
CASP5
|
caspase 5, apoptosis-related cysteine peptidase
|
|
chr3_+_130612806
|
0.044
|
NM_001199179
NM_001001485
NM_001199183
NM_001199184
NM_001199185
NM_014382
NM_001001486
NM_001001487
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chr1_+_26737306
|
0.044
|
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr12_-_122985396
|
0.044
|
|
ZCCHC8
|
zinc finger, CCHC domain containing 8
|
|
chr13_+_115079943
|
0.044
|
NM_001164144
NM_001164145
NM_032436
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1
|
|
chr19_+_57791281
|
0.044
|
|
ZNF460
|
zinc finger protein 460
|
|
chr8_+_124084936
|
0.043
|
|
WDR67
|
WD repeat domain 67
|
|
chr12_+_133657478
|
0.043
|
|
ZNF140
|
zinc finger protein 140
|
|
chr2_+_17935379
|
0.043
|
NM_001130009
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr12_-_11286842
|
0.043
|
NM_001097643
|
TAS2R30
|
taste receptor, type 2, member 30
|
|
chr2_-_74756973
|
0.043
|
NM_181575
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr3_+_154797644
|
0.043
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_+_63833324
|
0.043
|
|
ALG6
|
asparagine-linked glycosylation 6, alpha-1,3-glucosyltransferase homolog (S. cerevisiae)
|
|
chr9_-_86571652
|
0.043
|
NM_032307
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr11_+_118754474
|
0.043
|
NM_001716
|
CXCR5
|
chemokine (C-X-C motif) receptor 5
|
|
chr19_-_49250023
|
0.043
|
NM_182575
|
IZUMO1
|
izumo sperm-egg fusion 1
|
|
chr5_-_149792276
|
0.043
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr5_+_55149149
|
0.043
|
NM_001242636
NM_001242639
|
IL31RA
|
interleukin 31 receptor A
|
|
chr15_-_50411405
|
0.043
|
NM_024837
|
ATP8B4
|
ATPase, class I, type 8B, member 4
|
|
chr19_+_11200124
|
0.043
|
|
LDLR
|
low density lipoprotein receptor
|
|
chr1_+_63833260
|
0.042
|
NM_013339
|
ALG6
|
asparagine-linked glycosylation 6, alpha-1,3-glucosyltransferase homolog (S. cerevisiae)
|