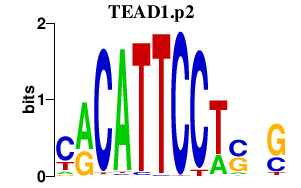
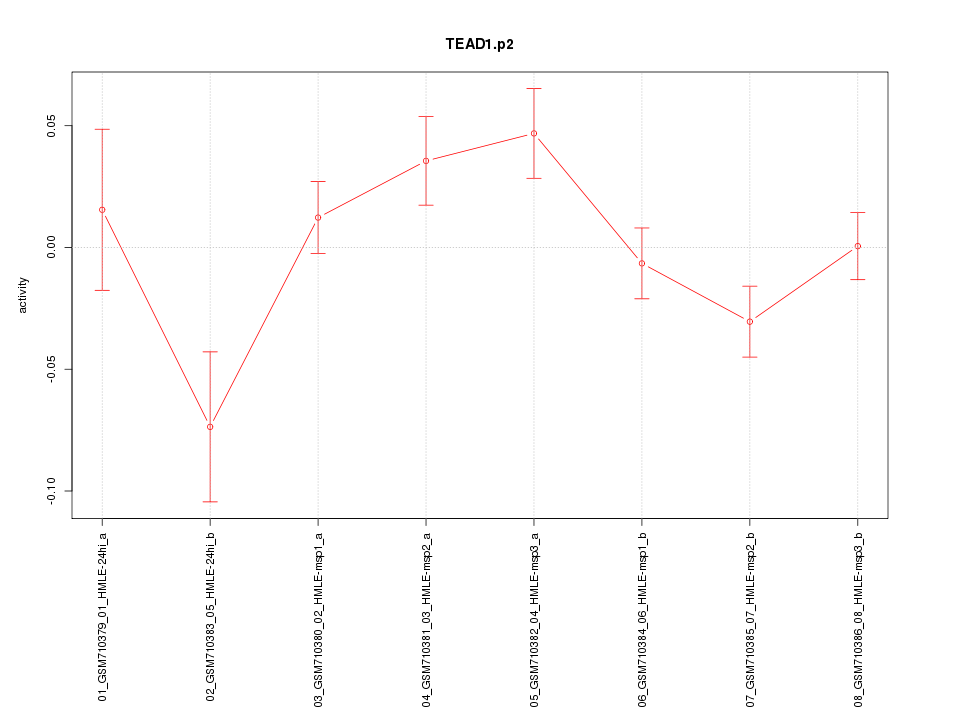
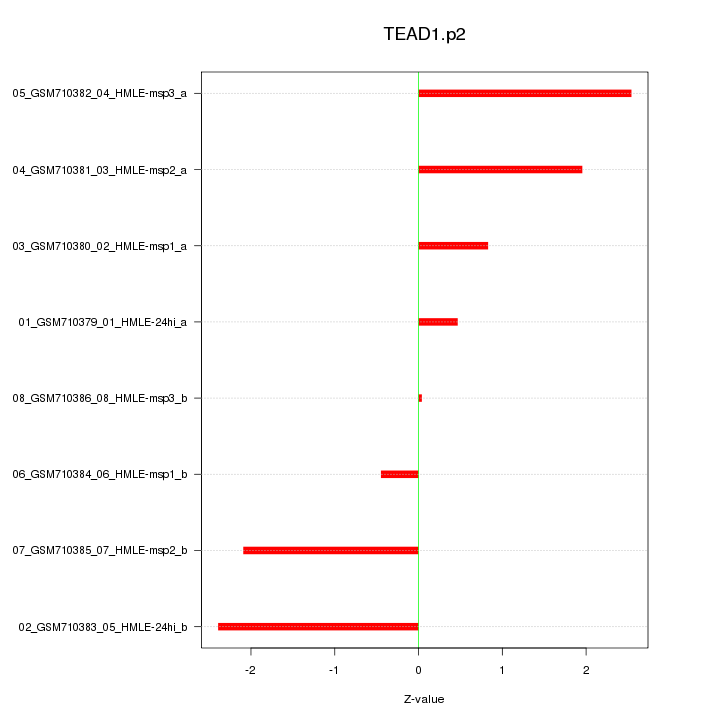
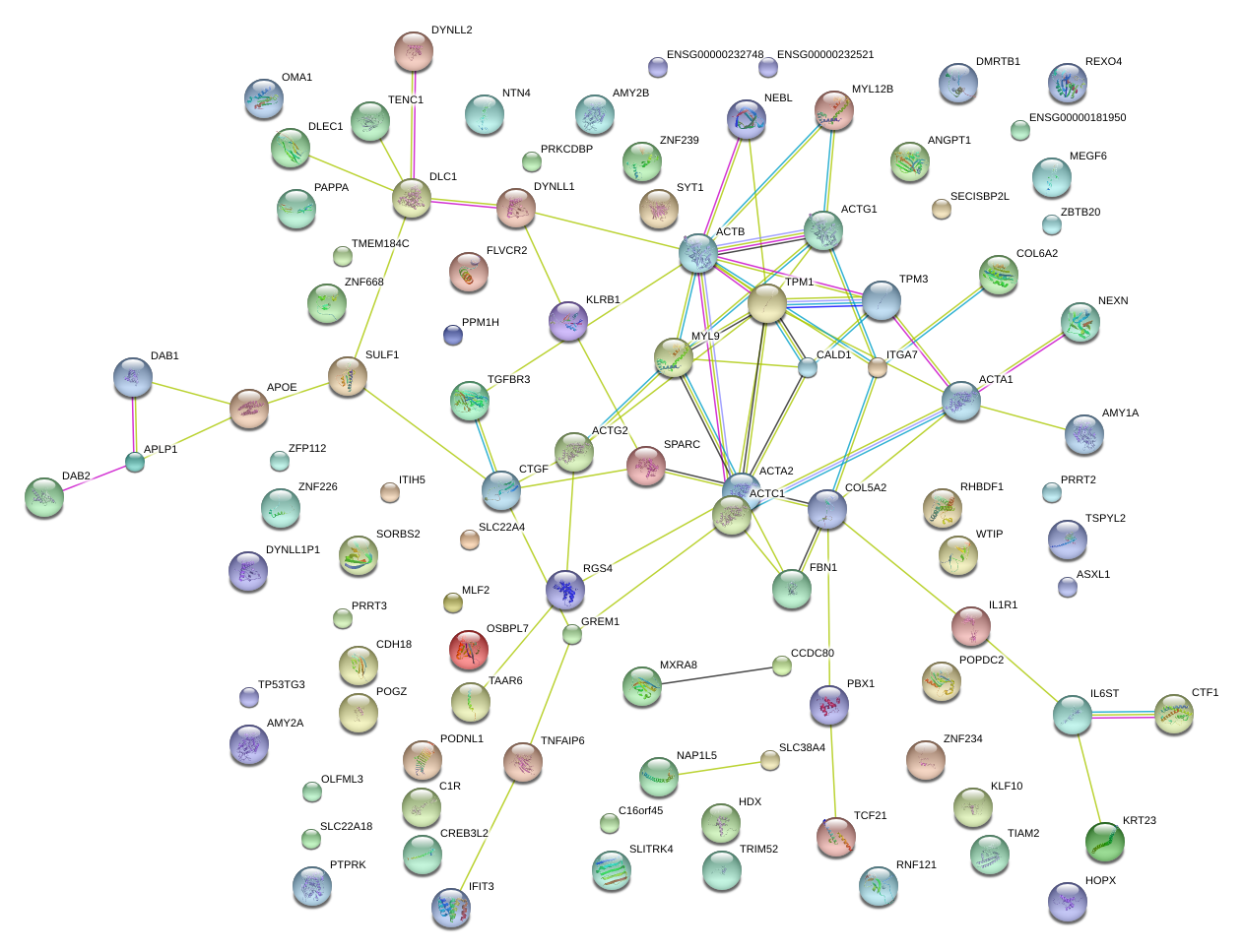
|
chr6_-_132272311
|
2.080
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr5_-_39424930
|
1.823
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr19_+_34972866
|
1.588
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr8_-_108510094
|
1.549
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr1_-_1293907
|
1.518
|
NM_032348
|
MXRA8
|
matrix-remodelling associated 8
|
|
chr15_-_48937795
|
1.376
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr12_-_47219734
|
1.303
|
NM_001143824
NM_018018
|
SLC38A4
|
solute carrier family 38, member 4
|
|
chr20_+_35169876
|
1.292
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr3_-_112359547
|
1.272
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_-_92351476
|
1.261
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr20_+_35169897
|
1.231
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr3_-_112359863
|
1.200
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr19_+_36359346
|
1.199
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr3_-_112359660
|
1.178
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112359962
|
1.169
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112359277
|
1.158
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112359565
|
1.144
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112359443
|
1.124
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112359251
|
1.105
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_114522029
|
1.091
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr1_-_3447974
|
1.084
|
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr5_-_39425030
|
1.067
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr5_-_19988293
|
1.049
|
NM_001167667
NM_004934
|
CDH18
|
cadherin 18, type 2
|
|
chr19_+_45409003
|
1.037
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr8_-_12973752
|
1.028
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr3_-_9994057
|
1.002
|
NM_207351
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr12_-_63328663
|
0.995
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr4_-_186733372
|
0.993
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr16_+_29823557
|
0.942
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr3_-_114343052
|
0.936
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_-_96184510
|
0.925
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr16_+_29823408
|
0.920
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr2_+_152214105
|
0.913
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr5_+_131630065
|
0.891
|
NM_003059
|
SLC22A4
|
solute carrier family 22 (organic cation/ergothioneine transporter), member 4
|
|
chr14_+_76044939
|
0.886
|
NM_017791
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr2_-_190044330
|
0.885
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr8_+_70378858
|
0.879
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr1_-_92351613
|
0.842
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr10_-_21186530
|
0.820
|
NM_006393
|
NEBL
|
nebulette
|
|
chr5_-_39425297
|
0.785
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr10_-_44063906
|
0.761
|
NM_005674
|
ZNF239
|
zinc finger protein 239
|
|
chr1_+_164528586
|
0.740
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr12_-_56101465
|
0.733
|
|
ITGA7
|
integrin, alpha 7
|
|
chr15_+_63335086
|
0.707
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr21_+_47545189
|
0.706
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr4_-_186578122
|
0.701
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr17_-_45899118
|
0.683
|
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr12_-_7245048
|
0.665
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr12_-_7245006
|
0.659
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr16_+_30907927
|
0.651
|
NM_001142544
NM_001330
|
CTF1
|
cardiotrophin 1
|
|
chr19_-_44831907
|
0.641
|
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr3_-_119379269
|
0.626
|
NM_022135
|
POPDC2
|
popeye domain containing 2
|
|
chr4_-_57547845
|
0.610
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr12_-_56101667
|
0.606
|
NM_001144996
NM_002206
|
ITGA7
|
integrin, alpha 7
|
|
chr17_-_45899133
|
0.604
|
NM_145798
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr1_+_78354199
|
0.603
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr16_+_33261514
|
0.596
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr12_+_53442732
|
0.594
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr4_-_89618928
|
0.588
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr15_+_63334753
|
0.588
|
NM_000366
NM_001018004
NM_001018005
NM_001018006
NM_001018007
NM_001018020
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr11_+_2924504
|
0.585
|
|
SLC22A18
|
solute carrier family 22, member 18
|
|
chr12_+_79258517
|
0.557
|
|
SYT1
|
synaptotagmin I
|
|
chr16_+_15528324
|
0.543
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr6_+_132891460
|
0.543
|
NM_175067
|
TAAR6
|
trace amine associated receptor 6
|
|
chr10_-_90712510
|
0.537
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr2_+_102759184
|
0.524
|
|
IL1R1
|
interleukin 1 receptor, type I
|
|
chr12_+_79258588
|
0.502
|
|
SYT1
|
synaptotagmin I
|
|
chr6_-_128841432
|
0.501
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr5_-_151066492
|
0.480
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr10_+_91087601
|
0.477
|
NM_001549
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr9_+_118916069
|
0.475
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr16_-_122573
|
0.467
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr19_+_44645728
|
0.456
|
|
ZNF234
|
zinc finger protein 234
|
|
chr12_+_79258448
|
0.453
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr6_+_155411422
|
0.450
|
NM_012454
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr12_-_63328857
|
0.448
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr5_-_180688095
|
0.441
|
NM_032765
|
TRIM52
|
tripartite motif containing 52
|
|
chrX_-_83757398
|
0.439
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr7_+_134464372
|
0.437
|
|
CALD1
|
caldesmon 1
|
|
chr1_-_58716207
|
0.435
|
NM_021080
|
DAB1
|
disabled homolog 1 (Drosophila)
|
|
chr1_+_104159953
|
0.427
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chr1_-_151431648
|
0.416
|
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chrX_+_53111482
|
0.414
|
NM_022117
|
TSPYL2
|
TSPY-like 2
|
|
chr6_+_134210264
|
0.412
|
|
TCF21
|
transcription factor 21
|
|
chr7_-_137686812
|
0.412
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr1_+_163038564
|
0.408
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr11_-_6341708
|
0.406
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr8_-_103666018
|
0.405
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chrX_-_142722869
|
0.405
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr11_+_71639767
|
0.397
|
NM_018320
|
RNF121
|
ring finger protein 121
|
|
chr5_-_55290601
|
0.396
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr19_-_14048877
|
0.392
|
|
PODNL1
|
podocan-like 1
|
|
chr17_-_39093671
|
0.391
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr12_+_79257772
|
0.382
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr19_-_14049167
|
0.380
|
NM_024825
|
PODNL1
|
podocan-like 1
|
|
chr20_+_30946786
|
0.376
|
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr1_+_53925071
|
0.368
|
NM_033067
|
DMRTB1
|
DMRT-like family B with proline-rich C-terminal, 1
|
|
chr4_+_148538564
|
0.367
|
|
TMEM184C
|
transmembrane protein 184C
|
|
chr16_-_31076408
|
0.362
|
NM_001172669
NM_001172670
|
ZNF668
|
zinc finger protein 668
|
|
chr10_-_7682741
|
0.360
|
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr15_+_33010174
|
0.357
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr17_-_4693595
|
0.357
|
|
LOC284014
|
uncharacterized LOC284014
|
|
chr15_-_49338496
|
0.355
|
NM_001193489
NM_014701
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chr19_+_50936159
|
0.355
|
NM_004533
|
MYBPC2
|
myosin binding protein C, fast type
|
|
chr18_+_3247825
|
0.354
|
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr17_-_63557764
|
0.354
|
|
AXIN2
|
axin 2
|
|
chr2_-_20212408
|
0.353
|
|
MATN3
|
matrilin 3
|
|
chr5_-_177423242
|
0.349
|
NM_006261
|
PROP1
|
PROP paired-like homeobox 1
|
|
chr7_-_137686778
|
0.343
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr9_-_73483933
|
0.339
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr1_+_163038934
|
0.339
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr11_+_5410606
|
0.335
|
NM_001004756
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1
|
|
chr19_-_9649257
|
0.334
|
NM_024106
|
ZNF426
|
zinc finger protein 426
|
|
chr19_-_13617037
|
0.331
|
NM_000068
NM_001127221
NM_001127222
NM_001174080
NM_023035
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr6_+_108881010
|
0.330
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr11_+_196760
|
0.322
|
NM_053280
|
ODF3
|
outer dense fiber of sperm tails 3
|
|
chr5_-_151066445
|
0.321
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr1_-_214724974
|
0.321
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr16_+_47495225
|
0.320
|
|
PHKB
|
phosphorylase kinase, beta
|
|
chr6_-_167571315
|
0.317
|
NM_005299
|
GPR31
|
G protein-coupled receptor 31
|
|
chr19_-_51472003
|
0.316
|
NM_001012964
NM_001012965
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr16_+_222845
|
0.310
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr3_-_46904879
|
0.310
|
NM_000258
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow
|
|
chr5_+_180688174
|
0.309
|
|
|
|
|
chr4_+_148538508
|
0.307
|
NM_018241
|
TMEM184C
|
transmembrane protein 184C
|
|
chr11_-_2170792
|
0.306
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr8_-_116681253
|
0.304
|
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chrX_+_107683015
|
0.304
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr17_-_63557615
|
0.302
|
NM_004655
|
AXIN2
|
axin 2
|
|
chr17_+_78194090
|
0.302
|
NM_001166347
NM_001166348
NM_001166349
NM_173626
|
SLC26A11
|
solute carrier family 26, member 11
|
|
chrX_+_86772714
|
0.302
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr8_-_95220769
|
0.299
|
NM_004063
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine)
|
|
chr17_-_48278987
|
0.298
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr5_-_38595506
|
0.297
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr9_-_95166827
|
0.296
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr1_+_226016428
|
0.294
|
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr6_+_31802644
|
0.291
|
NM_001040437
NM_001040438
|
C6orf48
|
chromosome 6 open reading frame 48
|
|
chr7_-_150946031
|
0.289
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr16_+_47495166
|
0.285
|
NM_000293
NM_001031835
|
PHKB
|
phosphorylase kinase, beta
|
|
chr22_+_24577443
|
0.285
|
NM_019601
|
SUSD2
|
sushi domain containing 2
|
|
chr16_+_1032063
|
0.284
|
|
SOX8
|
SRY (sex determining region Y)-box 8
|
|
chr2_-_158731622
|
0.275
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr6_+_52929795
|
0.275
|
NM_033481
|
FBXO9
|
F-box protein 9
|
|
chr6_+_84222252
|
0.273
|
|
PRSS35
|
protease, serine, 35
|
|
chr11_+_48002063
|
0.272
|
NM_001098503
NM_002843
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr19_+_14491920
|
0.270
|
NM_001025160
NM_001784
NM_078481
|
CD97
|
CD97 molecule
|
|
chr16_+_226678
|
0.270
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr5_-_95158365
|
0.269
|
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr11_+_48002120
|
0.268
|
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr19_+_12075868
|
0.268
|
NM_001012753
|
ZNF763
|
zinc finger protein 763
|
|
chr2_-_20212448
|
0.265
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr3_+_38347426
|
0.264
|
NM_004803
|
SLC22A14
|
solute carrier family 22, member 14
|
|
chr7_+_8008416
|
0.263
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr9_+_137533713
|
0.263
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr1_-_17307107
|
0.262
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr8_-_38325241
|
0.261
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chrX_+_79675945
|
0.259
|
NM_152630
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chr12_-_122240827
|
0.258
|
|
|
|
|
chr2_+_112895958
|
0.258
|
NM_001128165
NM_153214
|
FBLN7
|
fibulin 7
|
|
chr14_-_89020888
|
0.256
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr7_-_137686814
|
0.256
|
NM_001253775
NM_194071
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr1_-_198510074
|
0.255
|
NM_133262
NM_133326
|
ATP6V1G3
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3
|
|
chr5_-_43412417
|
0.253
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr5_+_140749830
|
0.251
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr17_-_48278874
|
0.250
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr8_+_1772095
|
0.249
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr6_+_52930160
|
0.245
|
|
FBXO9
|
F-box protein 9
|
|
chr16_-_47495095
|
0.244
|
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1
|
|
chr19_+_11998609
|
0.239
|
NM_021915
|
ZNF69
|
zinc finger protein 69
|
|
chr16_+_28889808
|
0.239
|
NM_004320
NM_173201
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr6_-_75915753
|
0.235
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr5_+_140868721
|
0.235
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr14_-_77494621
|
0.235
|
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like
|
|
chr15_-_69707358
|
0.235
|
|
LOC145694
|
uncharacterized LOC145694
|
|
chr14_+_21467413
|
0.234
|
NM_014579
|
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2
|
|
chr9_+_34990715
|
0.234
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr22_-_36635999
|
0.232
|
NM_145637
|
APOL2
|
apolipoprotein L, 2
|
|
chr3_+_9944511
|
0.232
|
NM_001193380
NM_153480
|
IL17RE
|
interleukin 17 receptor E
|
|
chr6_-_83903632
|
0.230
|
NM_001199917
|
PGM3
|
phosphoglucomutase 3
|
|
chr6_-_31681845
|
0.229
|
|
|
|
|
chr1_+_51435911
|
0.229
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr11_-_6643809
|
0.228
|
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr2_-_96482382
|
0.225
|
|
LINC00342
|
long intergenic non-protein coding RNA 342
|
|
chr8_+_145086444
|
0.225
|
NM_001134374
NM_198572
|
SPATC1
|
spermatogenesis and centriole associated 1
|
|
chr1_+_218518675
|
0.224
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr9_-_130533509
|
0.224
|
NM_001252334
NM_005489
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr1_-_162381805
|
0.223
|
NM_053282
|
SH2D1B
|
SH2 domain containing 1B
|
|
chr16_+_33204979
|
0.223
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr1_-_120935893
|
0.223
|
|
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64)
|
|
chr3_-_149086884
|
0.223
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr3_-_127541160
|
0.222
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr6_+_52930222
|
0.220
|
NM_033480
|
FBXO9
|
F-box protein 9
|
|
chr5_-_150138081
|
0.217
|
NM_001135644
|
DCTN4
|
dynactin 4 (p62)
|
|
chr5_-_95158465
|
0.214
|
NM_001118890
NM_001243658
NM_001243659
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr2_-_217540722
|
0.213
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr14_-_35591151
|
0.209
|
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr6_+_83903031
|
0.208
|
NM_033411
|
RWDD2A
|
RWD domain containing 2A
|
|
chr11_+_22689653
|
0.207
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr6_+_52930217
|
0.206
|
|
FBXO9
|
F-box protein 9
|
|
chr19_-_49371710
|
0.206
|
NM_001161354
NM_020904
|
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4
|