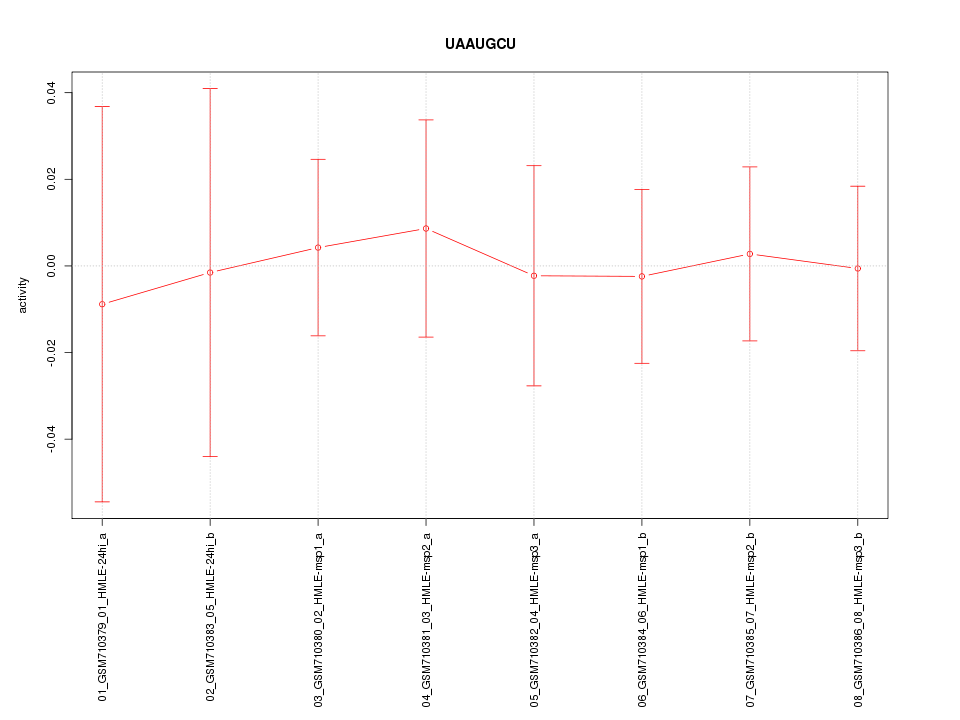
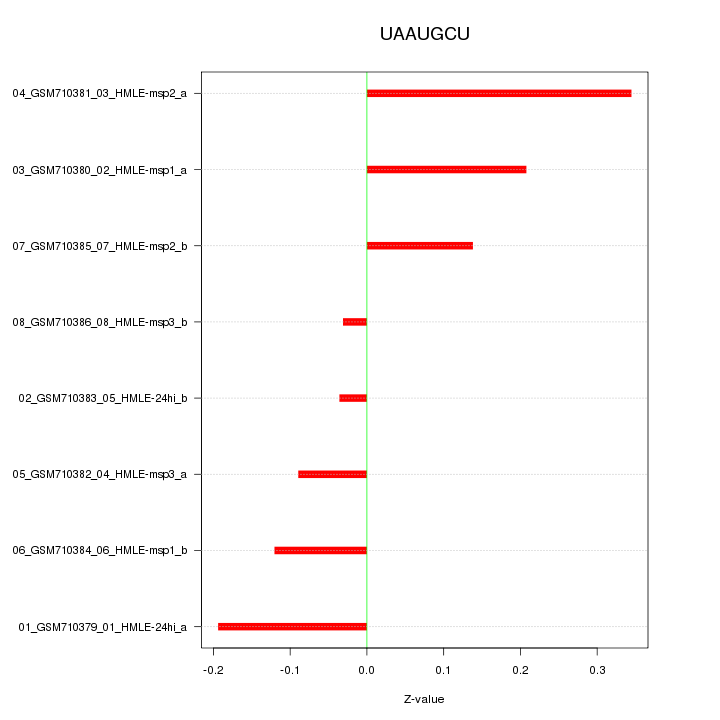
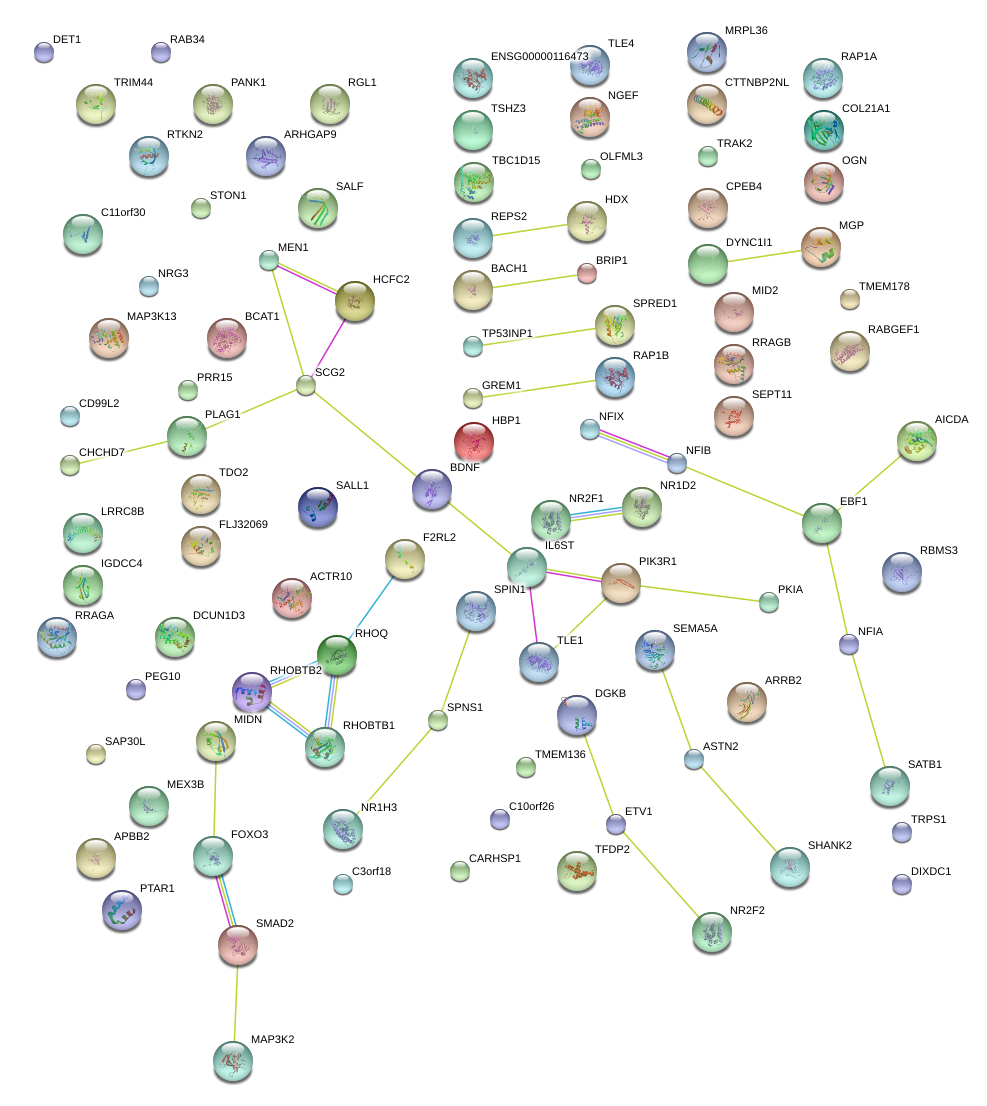
|
chr1_+_114522029
|
0.133
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr1_+_183605207
|
0.090
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr5_-_158526698
|
0.085
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chrX_+_16964730
|
0.077
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr8_-_95961543
|
0.076
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr12_-_15038724
|
0.069
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr10_+_83637442
|
0.066
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chrX_-_150067127
|
0.064
|
NM_001184808
NM_001242614
NM_031462
NM_134445
NM_134446
|
CD99L2
|
CD99 molecule-like 2
|
|
chr10_+_83635069
|
0.064
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr11_-_27743604
|
0.061
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27681195
|
0.059
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27721179
|
0.058
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr8_-_116681103
|
0.058
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr15_-_89089792
|
0.057
|
NM_001144074
NM_017996
|
DET1
|
de-etiolated homolog 1 (Arabidopsis)
|
|
chr11_+_47279467
|
0.057
|
NM_001130101
NM_005693
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr2_+_46769821
|
0.057
|
NM_012249
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr11_-_27722599
|
0.056
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27721975
|
0.055
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27722446
|
0.055
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_-_9546157
|
0.055
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr11_-_70935807
|
0.054
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr11_-_70507911
|
0.053
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr11_+_47270433
|
0.053
|
NM_001130102
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr11_-_27741192
|
0.051
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr16_-_51184507
|
0.051
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr11_-_27742224
|
0.049
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27723061
|
0.049
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr9_-_95166827
|
0.048
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr4_+_77870864
|
0.047
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr15_-_82338348
|
0.047
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr11_+_120195837
|
0.043
|
NM_001198670
NM_001198671
NM_001198672
NM_001198673
NM_001198674
NM_001198675
NM_174926
|
TMEM136
|
transmembrane protein 136
|
|
chr16_-_51185150
|
0.043
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr3_+_29322802
|
0.042
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr6_+_108882068
|
0.041
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr3_+_185000714
|
0.040
|
NM_001242314
NM_001242317
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr3_-_18466759
|
0.039
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr6_+_108881010
|
0.039
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr19_+_1248548
|
0.039
|
NM_177401
|
MIDN
|
midnolin
|
|
chr3_-_18480203
|
0.038
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr8_+_79428335
|
0.038
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chrX_+_55744049
|
0.037
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr17_+_4613780
|
0.037
|
NM_004313
NM_199004
|
ARRB2
|
arrestin, beta 2
|
|
chr19_-_31840118
|
0.035
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr15_+_33010174
|
0.035
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr7_+_66093854
|
0.035
|
NM_001167961
NM_153033
|
KCTD7
|
potassium channel tetramerisation domain containing 7
|
|
chr11_+_35684342
|
0.034
|
NM_017583
|
TRIM44
|
tripartite motif containing 44
|
|
chr17_-_27045172
|
0.033
|
NM_001142624
NM_001142625
NM_001144943
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr10_-_91403630
|
0.032
|
NM_138316
NM_148978
|
PANK1
|
pantothenate kinase 1
|
|
chr2_-_202316227
|
0.032
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr10_-_91405320
|
0.032
|
NM_148977
|
PANK1
|
pantothenate kinase 1
|
|
chr5_+_153825497
|
0.030
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr17_-_27044728
|
0.030
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr16_-_8962842
|
0.030
|
NM_014316
|
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa
|
|
chr2_-_233877920
|
0.029
|
NM_019850
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr21_+_30671736
|
0.029
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr16_-_8962247
|
0.029
|
NM_001042476
|
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa
|
|
chr7_+_29603388
|
0.029
|
NM_175887
|
PRR15
|
proline rich 15
|
|
chr9_+_82186846
|
0.028
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr3_-_141719188
|
0.028
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr5_+_67511578
|
0.027
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr5_+_67584251
|
0.027
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chrX_+_107069081
|
0.027
|
NM_012216
NM_052817
|
MID2
|
midline 2
|
|
chr2_+_39892637
|
0.027
|
NM_001167959
|
TMEM178
|
transmembrane protein 178
|
|
chr3_-_141868208
|
0.027
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr5_+_67588395
|
0.027
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr15_+_38544966
|
0.027
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr7_-_14031049
|
0.026
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr7_-_14025826
|
0.026
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr5_+_67586466
|
0.026
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr2_+_39893021
|
0.026
|
NM_152390
|
TMEM178
|
transmembrane protein 178
|
|
chr8_-_57123858
|
0.026
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr7_-_14030864
|
0.026
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr7_-_14029641
|
0.025
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr8_+_57124314
|
0.025
|
NM_001011667
NM_001011668
NM_001011669
NM_001011670
NM_001011671
NM_024300
|
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7
|
|
chr9_-_120177307
|
0.024
|
NM_014010
|
ASTN2
|
astrotactin 2
|
|
chr2_+_48757292
|
0.024
|
NM_001198595
NM_006873
|
STON1
|
stonin 1
|
|
chr16_-_20911560
|
0.024
|
NM_173475
|
DCUN1D3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chrX_-_83757398
|
0.023
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr10_+_104535887
|
0.023
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr3_-_141747434
|
0.023
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr12_-_8765438
|
0.023
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr7_+_106809405
|
0.022
|
NM_001244262
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr2_-_224467142
|
0.022
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr5_-_75919051
|
0.022
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr1_+_61547625
|
0.022
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr10_+_104503724
|
0.021
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr15_+_96875793
|
0.021
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_+_89990396
|
0.021
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr12_+_72233486
|
0.021
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr4_-_40859081
|
0.021
|
NM_001166051
NM_001166052
NM_001166053
NM_001166054
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr5_-_55290601
|
0.021
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr12_+_104458235
|
0.020
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr9_+_91003238
|
0.020
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr18_-_45457454
|
0.020
|
NM_001003652
NM_001135937
|
SMAD2
|
SMAD family member 2
|
|
chr4_-_41216455
|
0.019
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr9_-_72374855
|
0.019
|
NM_001099666
|
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1
|
|
chr21_+_30671217
|
0.019
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr12_-_25055228
|
0.019
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr7_+_95401817
|
0.019
|
NM_001135556
NM_001135557
NM_004411
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1
|
|
chr12_-_25055966
|
0.018
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr1_+_61542928
|
0.018
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr3_+_23987514
|
0.018
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr2_-_233792825
|
0.018
|
NM_001114090
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr1_+_90024271
|
0.018
|
NM_015350
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr6_-_56112315
|
0.018
|
NM_030820
|
COL21A1
|
collagen, type XXI, alpha 1
|
|
chr3_-_50608421
|
0.018
|
NM_001171741
|
C3orf18
|
chromosome 3 open reading frame 18
|
|
chr18_-_45456925
|
0.018
|
NM_005901
|
SMAD2
|
SMAD family member 2
|
|
chr15_-_65715385
|
0.018
|
NM_020962
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr10_-_64028457
|
0.018
|
NM_145307
|
RTKN2
|
rhotekin 2
|
|
chr10_-_62703822
|
0.017
|
NM_001242359
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr11_+_111848008
|
0.017
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|
|
chr7_-_14880954
|
0.017
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr12_-_25102281
|
0.017
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr15_+_96869156
|
0.017
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr2_-_128100673
|
0.017
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr7_+_94285676
|
0.017
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr7_+_94285620
|
0.017
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr1_+_112938799
|
0.017
|
NM_018704
|
CTTNBP2NL
|
CTTNBP2 N-terminal like
|
|
chr14_+_58666818
|
0.017
|
NM_018477
|
ACTR10
|
actin-related protein 10 homolog (S. cerevisiae)
|
|
chr3_+_23986735
|
0.016
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_-_50605062
|
0.016
|
NM_001171740
NM_001171743
NM_016210
|
C3orf18
|
chromosome 3 open reading frame 18
|
|
chr11_+_76156059
|
0.016
|
NM_020193
|
C11orf30
|
chromosome 11 open reading frame 30
|
|
chr12_+_69004614
|
0.016
|
NM_001010942
NM_001251917
NM_001251918
NM_001251921
NM_001251922
NM_015646
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr5_+_173315313
|
0.016
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr15_+_96873845
|
0.016
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr5_+_161277702
|
0.016
|
NM_001127648
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_-_225840660
|
0.015
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr4_+_129730772
|
0.015
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chr5_+_161277391
|
0.015
|
NM_001127647
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr10_+_114206708
|
0.015
|
NM_145206
|
VTI1A
|
vesicle transport through interaction with t-SNAREs homolog 1A (yeast)
|
|
chr5_+_161274939
|
0.015
|
NM_001127644
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_+_154377668
|
0.015
|
NM_000565
NM_001206866
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr15_+_96876568
|
0.015
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_-_91487769
|
0.015
|
NM_032186
NM_201269
|
ZNF644
|
zinc finger protein 644
|
|
chrX_+_136648313
|
0.015
|
NM_003413
|
ZIC3
|
Zic family member 3
|
|
chr5_+_161275541
|
0.015
|
NM_001127645
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_+_109102970
|
0.014
|
NM_001010883
|
FAM102B
|
family with sequence similarity 102, member B
|
|
chr10_+_181410
|
0.014
|
NM_001202464
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr11_+_111807857
|
0.014
|
NM_001037954
|
DIXDC1
|
DIX domain containing 1
|
|
chr1_-_33815498
|
0.014
|
NM_004427
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr1_-_23857711
|
0.014
|
NM_004091
|
E2F2
|
E2F transcription factor 2
|
|
chr5_+_161274684
|
0.014
|
NM_001127643
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chrX_-_19988381
|
0.014
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr3_+_187930720
|
0.014
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr10_+_86088156
|
0.014
|
NM_018999
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr9_-_15510209
|
0.014
|
NM_021144
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr20_+_11898536
|
0.014
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr15_+_41186601
|
0.014
|
NM_020857
|
VPS18
|
vacuolar protein sorting 18 homolog (S. cerevisiae)
|
|
chr5_+_135468522
|
0.014
|
NM_001001419
NM_001001420
NM_005903
|
SMAD5
|
SMAD family member 5
|
|
chr6_+_158589378
|
0.014
|
NM_207118
|
GTF2H5
|
general transcription factor IIH, polypeptide 5
|
|
chr17_-_67323185
|
0.014
|
NM_172232
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chrX_-_48937560
|
0.014
|
NM_001029896
|
WDR45
|
WD repeat domain 45
|
|
chr1_+_110036657
|
0.014
|
NM_001134402
NM_001134400
NM_001134403
NM_001134404
NM_182580
|
CYB561D1
|
cytochrome b-561 domain containing 1
|
|
chr2_-_180129245
|
0.013
|
NM_178123
|
SESTD1
|
SEC14 and spectrin domains 1
|
|
chr17_-_71228482
|
0.013
|
NM_001098832
NM_032837
|
FAM104A
|
family with sequence similarity 104, member A
|
|
chr19_+_18111923
|
0.013
|
NM_001025604
|
ARRDC2
|
arrestin domain containing 2
|
|
chr19_+_18118921
|
0.013
|
NM_015683
|
ARRDC2
|
arrestin domain containing 2
|
|
chr17_+_66508525
|
0.013
|
NM_002734
NM_212472
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr5_+_161275897
|
0.013
|
NM_001127646
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr9_+_79056581
|
0.013
|
NM_001097634
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr1_-_108231122
|
0.013
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr20_+_60813577
|
0.013
|
NM_014835
NM_144498
|
OSBPL2
|
oxysterol binding protein-like 2
|
|
chr3_+_141043054
|
0.013
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr12_+_27397077
|
0.012
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr2_-_200335988
|
0.012
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr2_-_200329810
|
0.012
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr12_+_93771607
|
0.012
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr7_+_117824085
|
0.012
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr3_+_187943192
|
0.012
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr19_-_1605464
|
0.012
|
NM_006830
|
UQCR11
|
ubiquinol-cytochrome c reductase, complex III subunit XI
|
|
chr1_-_52456292
|
0.012
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr1_+_160175114
|
0.012
|
NM_003768
|
PEA15
|
phosphoprotein enriched in astrocytes 15
|
|
chr20_+_48807119
|
0.012
|
NM_005194
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr2_+_201997687
|
0.012
|
NM_001202518
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr3_+_187871473
|
0.012
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_+_185080835
|
0.012
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr10_+_180404
|
0.012
|
NM_006624
NM_001202465
NM_001202466
NM_212479
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr2_+_205410465
|
0.012
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr9_+_79115548
|
0.011
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr17_-_56065596
|
0.011
|
NM_007146
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr6_+_149639435
|
0.011
|
NM_015093
|
TAB2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2
|
|
chr17_-_44302716
|
0.011
|
NM_001193465
|
KIAA1267
|
KIAA1267
|
|
chr2_+_208394615
|
0.011
|
NM_004379
NM_134442
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr1_-_108507474
|
0.011
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chrX_-_153979161
|
0.011
|
NM_001081573
NM_080612
|
GAB3
|
GRB2-associated binding protein 3
|
|
chr17_-_4167028
|
0.011
|
NM_016376
NM_020740
|
ANKFY1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr9_+_79074067
|
0.011
|
NM_001097633
NM_001490
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr9_-_14398981
|
0.011
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr9_+_79093256
|
0.011
|
NM_001097635
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr1_+_61547388
|
0.011
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr20_+_11871364
|
0.011
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr2_-_200322699
|
0.011
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr1_-_25573933
|
0.010
|
NM_020317
|
C1orf63
|
chromosome 1 open reading frame 63
|
|
chr15_-_60919642
|
0.010
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr10_+_74927865
|
0.010
|
NM_173348
|
FAM149B1
|
family with sequence similarity 149, member B1
|
|
chr9_-_119449374
|
0.010
|
NM_198186
NM_001184734
NM_001184735
NM_198187
NM_198188
|
ASTN2
|
astrotactin 2
|
|
chr17_+_66508098
|
0.010
|
NM_212471
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr19_+_34663311
|
0.010
|
NM_001114093
NM_015578
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr17_+_57642885
|
0.010
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr1_-_229569839
|
0.010
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|