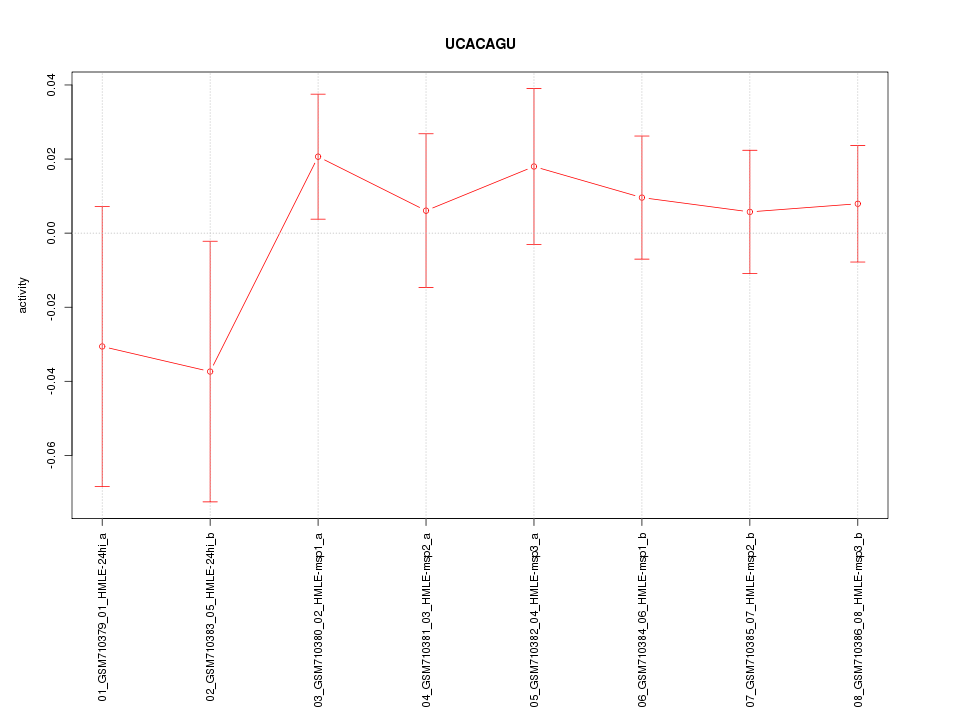
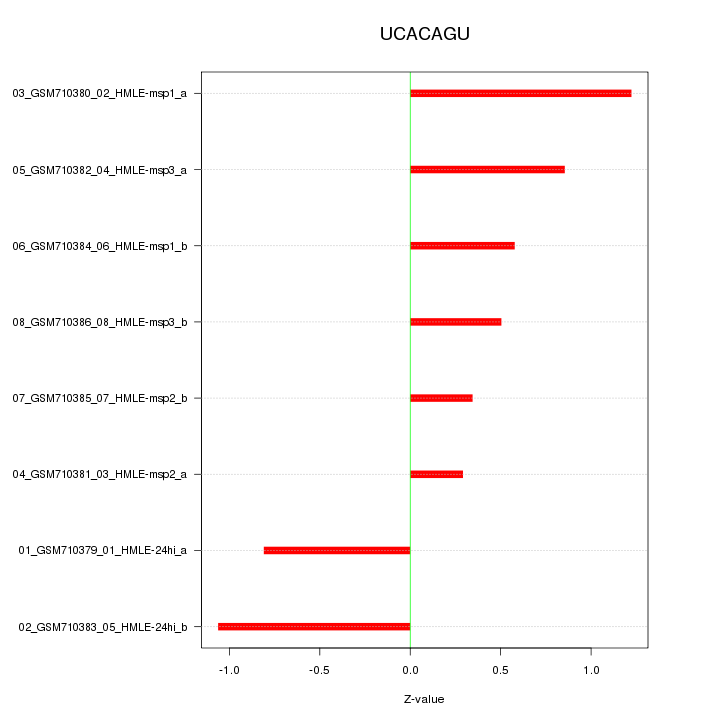
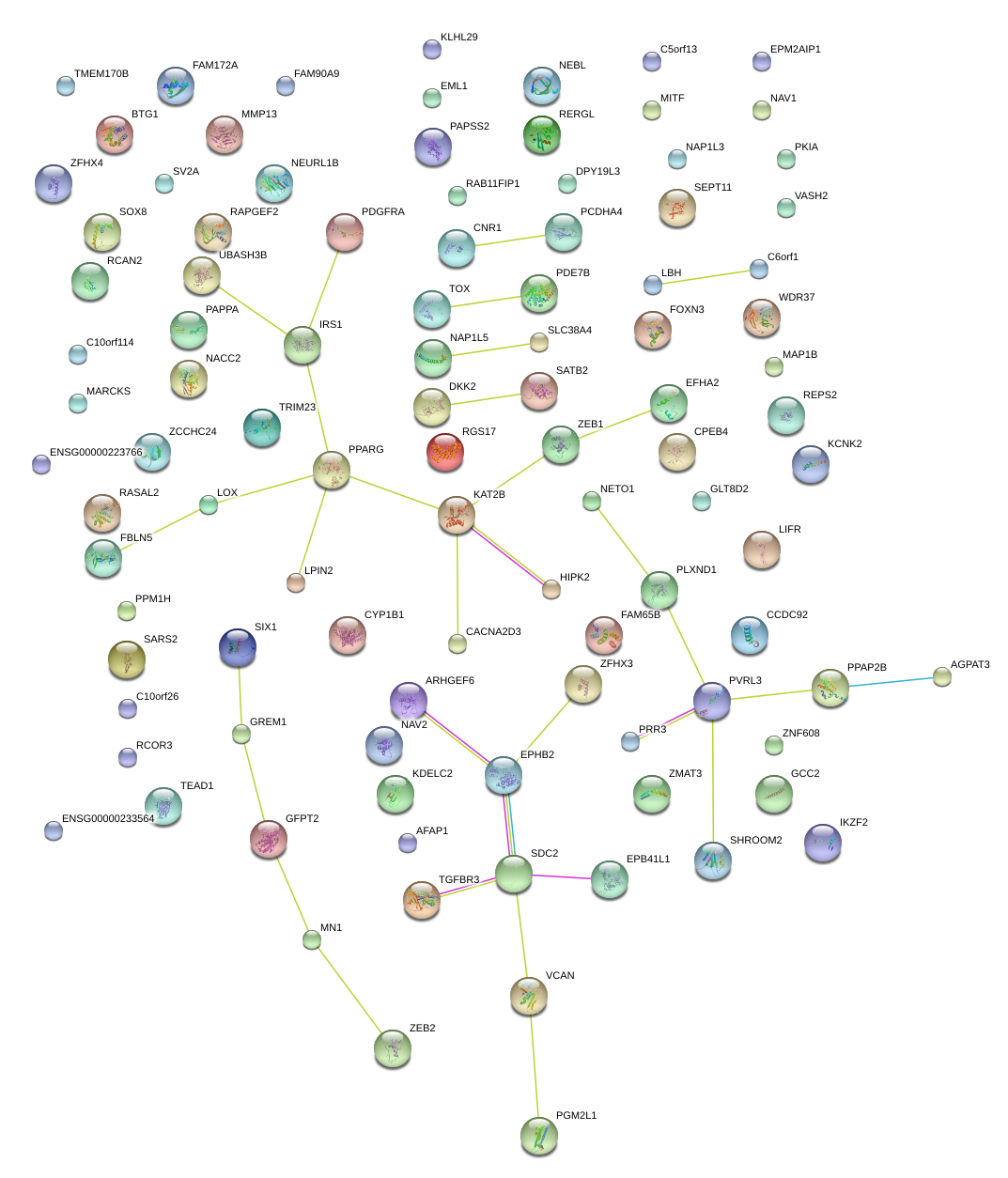
|
chr14_-_92414004
|
1.400
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr15_+_33010174
|
1.360
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr5_-_111093251
|
1.345
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093030
|
1.333
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111312522
|
1.333
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111092918
|
1.323
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_-_92351613
|
1.309
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr5_-_111093792
|
1.305
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_-_92371558
|
1.290
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_-_57045235
|
1.201
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr10_-_21463019
|
1.168
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr6_-_46293628
|
1.109
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr6_-_88875766
|
1.025
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr5_+_172068188
|
0.927
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr5_+_71403040
|
0.916
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr3_+_12392993
|
0.886
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr3_+_12330336
|
0.880
|
NM_138711
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr10_-_21186530
|
0.865
|
NM_006393
|
NEBL
|
nebulette
|
|
chr3_+_12329332
|
0.852
|
NM_005037
NM_138712
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr5_-_179780314
|
0.846
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chrX_+_16964730
|
0.827
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr10_-_81205091
|
0.821
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr2_-_38303307
|
0.774
|
NM_000104
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr5_-_111091947
|
0.759
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_215178884
|
0.718
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr1_+_215256559
|
0.713
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr8_+_79428335
|
0.687
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr11_+_19734821
|
0.682
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr5_-_121412805
|
0.670
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr5_-_121414011
|
0.657
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr12_-_47219734
|
0.653
|
NM_001143824
NM_018018
|
SLC38A4
|
solute carrier family 38, member 4
|
|
chr4_+_55095263
|
0.646
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr3_-_129325175
|
0.645
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr11_+_19372270
|
0.637
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr11_-_74109417
|
0.637
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr2_-_200335988
|
0.632
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr14_+_100259445
|
0.620
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr2_-_200329810
|
0.610
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr14_-_61115906
|
0.602
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr2_-_200322699
|
0.596
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr6_+_30524485
|
0.587
|
NM_001077497
NM_025263
|
PRR3
|
proline rich 3
|
|
chr8_-_60031545
|
0.581
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr12_-_124457099
|
0.574
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr4_-_89618928
|
0.572
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr4_-_107957452
|
0.569
|
NM_014421
|
DKK2
|
dickkopf 2 homolog (Xenopus laevis)
|
|
chr7_-_139477437
|
0.568
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr2_-_214016332
|
0.565
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr14_-_89883306
|
0.561
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr6_+_136172802
|
0.542
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr2_+_23608297
|
0.528
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chr6_-_153452383
|
0.526
|
NM_012419
|
RGS17
|
regulator of G-protein signaling 17
|
|
chr2_+_30454396
|
0.510
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr14_-_90085325
|
0.476
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr8_+_16884745
|
0.470
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr6_-_88854989
|
0.469
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr16_+_1031807
|
0.468
|
NM_014587
|
SOX8
|
SRY (sex determining region Y)-box 8
|
|
chr5_-_124080773
|
0.452
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr6_+_11538486
|
0.445
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr18_-_3011944
|
0.444
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr1_+_213123853
|
0.443
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr20_+_34700257
|
0.441
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr8_+_77593506
|
0.439
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr4_+_160188997
|
0.438
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr16_-_73082169
|
0.433
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr8_+_97505876
|
0.423
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr1_+_201708940
|
0.418
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr20_+_34742656
|
0.416
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr5_-_38556682
|
0.403
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr12_-_63328663
|
0.389
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr5_-_38595506
|
0.384
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr3_-_178789597
|
0.382
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr5_+_82767492
|
0.382
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr1_+_201617449
|
0.381
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr1_-_149889362
|
0.376
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chrX_+_9754465
|
0.373
|
NM_001649
|
SHROOM2
|
shroom family member 2
|
|
chr2_+_109065576
|
0.371
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr22_-_28197469
|
0.369
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr3_+_69812961
|
0.369
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_+_77870864
|
0.369
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr3_+_69812620
|
0.368
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_69915374
|
0.367
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr11_-_108368918
|
0.366
|
NM_153705
|
KDELC2
|
KDEL (Lys-Asp-Glu-Leu) containing 2
|
|
chr1_+_23037329
|
0.364
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr3_+_69788562
|
0.361
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_20081523
|
0.357
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr2_-_214015053
|
0.356
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chrX_-_92928566
|
0.343
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr9_-_138987096
|
0.339
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr10_-_21786205
|
0.339
|
NM_001010911
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr19_+_32896515
|
0.336
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr11_-_102826433
|
0.335
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr19_+_32897021
|
0.333
|
NM_207325
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr21_+_45285101
|
0.330
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr10_+_1102741
|
0.329
|
NM_014023
|
WDR37
|
WD repeat domain 37
|
|
chr5_-_64920149
|
0.329
|
NM_001656
NM_033227
NM_033228
|
TRIM23
|
tripartite motif containing 23
|
|
chr1_+_211433285
|
0.322
|
NM_018254
|
RCOR3
|
REST corepressor 3
|
|
chr2_-_145277879
|
0.321
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr10_+_89419352
|
0.319
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr21_+_45345278
|
0.314
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr11_+_122526339
|
0.313
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chrX_-_135863502
|
0.312
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr9_+_118916069
|
0.309
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr4_-_7941092
|
0.309
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr18_-_70534809
|
0.306
|
NM_001201465
NM_138966
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr5_+_173315313
|
0.305
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr2_-_227663454
|
0.303
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr3_+_54156691
|
0.299
|
NM_018398
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr11_+_12695851
|
0.296
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr10_+_104503724
|
0.293
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr6_-_24911194
|
0.290
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr6_+_114178516
|
0.289
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr5_-_93447237
|
0.285
|
NM_001163417
NM_001163418
NM_032042
|
FAM172A
|
family with sequence similarity 172, member A
|
|
chr10_+_104535887
|
0.284
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr3_+_119187782
|
0.283
|
NM_152305
|
POGLUT1
|
protein O-glucosyltransferase 1
|
|
chr8_-_37756971
|
0.281
|
NM_001002814
NM_025151
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr12_-_92539614
|
0.280
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr1_+_178310605
|
0.280
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr7_+_90338711
|
0.279
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr12_+_79257772
|
0.276
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr12_-_49182718
|
0.275
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr14_+_51955854
|
0.274
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr15_-_37390372
|
0.272
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr21_-_34100228
|
0.272
|
NM_001160302
NM_001160306
|
SYNJ1
|
synaptojanin 1
|
|
chr7_-_28998028
|
0.271
|
NM_014817
|
TRIL
|
TLR4 interactor with leucine-rich repeats
|
|
chr6_-_53530505
|
0.269
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr2_-_85555343
|
0.268
|
NM_001206840
NM_001206841
NM_001206844
NM_006464
|
TGOLN2
|
trans-golgi network protein 2
|
|
chr12_+_54422193
|
0.266
|
NM_004503
|
HOXC6
|
homeobox C6
|
|
chr2_+_43864433
|
0.262
|
NM_172069
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr19_-_7293876
|
0.260
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr2_+_24807345
|
0.260
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr12_+_103981050
|
0.258
|
NM_017564
|
STAB2
|
stabilin 2
|
|
chr3_+_69985750
|
0.257
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_+_38261079
|
0.255
|
NM_152496
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chrX_+_86772714
|
0.254
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr3_+_43732358
|
0.253
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr12_+_79439432
|
0.250
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr2_-_192711923
|
0.248
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr9_+_18474078
|
0.247
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr12_-_110318178
|
0.246
|
NM_016433
|
GLTP
|
glycolipid transfer protein
|
|
chr20_-_3149070
|
0.245
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr7_-_27183225
|
0.243
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr19_+_15218141
|
0.243
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr7_+_7222142
|
0.243
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr14_+_72398816
|
0.242
|
NM_001204423
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr8_-_93029864
|
0.238
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_+_11886739
|
0.235
|
NM_145693
|
LPIN1
|
lipin 1
|
|
chr14_+_52118539
|
0.234
|
NM_152330
|
FRMD6
|
FERM domain containing 6
|
|
chr8_-_93115453
|
0.233
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr21_-_34100316
|
0.232
|
NM_003895
NM_203446
|
SYNJ1
|
synaptojanin 1
|
|
chr14_+_55034329
|
0.230
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr6_+_126102278
|
0.230
|
NM_001199619
NM_001199620
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr5_+_72143929
|
0.228
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr15_+_91447419
|
0.228
|
NM_006122
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2
|
|
chr20_-_48732479
|
0.227
|
NM_022442
NM_021988
NM_199144
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr12_-_53893195
|
0.227
|
NM_001193511
NM_006301
|
MAP3K12
|
mitogen-activated protein kinase kinase kinase 12
|
|
chrX_+_102632108
|
0.227
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr12_+_79258448
|
0.226
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr22_-_44708730
|
0.225
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr2_+_202899309
|
0.224
|
NM_003507
|
FZD7
|
frizzled family receptor 7
|
|
chr1_-_47134039
|
0.221
|
NM_001042546
NM_022745
|
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1
|
|
chr1_+_38259458
|
0.220
|
NM_001031740
NM_001113482
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr20_+_56964162
|
0.219
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr16_+_66878281
|
0.219
|
NM_005182
|
CA7
|
carbonic anhydrase VII
|
|
chr5_-_132299263
|
0.217
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr12_-_90049818
|
0.216
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr15_+_96875793
|
0.214
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chrX_+_102631267
|
0.213
|
NM_206915
NM_206917
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr1_-_225840660
|
0.212
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr1_+_178062829
|
0.211
|
NM_170692
|
RASAL2
|
RAS protein activator like 2
|
|
chr5_+_176560627
|
0.210
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr1_-_21605984
|
0.209
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr16_-_25268841
|
0.208
|
NM_001012981
|
ZKSCAN2
|
zinc finger with KRAB and SCAN domains 2
|
|
chr1_-_95392501
|
0.207
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr12_-_31743822
|
0.205
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr16_+_66878840
|
0.205
|
NM_001014435
|
CA7
|
carbonic anhydrase VII
|
|
chr17_-_58603492
|
0.205
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chrX_-_39956655
|
0.203
|
NM_001123385
NM_017745
|
BCOR
|
BCL6 corepressor
|
|
chr4_+_148402068
|
0.201
|
NM_001166055
NM_001957
|
EDNRA
|
endothelin receptor type A
|
|
chr16_-_73092533
|
0.199
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr12_+_12764755
|
0.198
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr1_-_21616648
|
0.198
|
NM_001113349
|
ECE1
|
endothelin converting enzyme 1
|
|
chr3_+_142838596
|
0.197
|
NM_004267
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2
|
|
chr3_-_179169330
|
0.197
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr1_+_211432693
|
0.196
|
NM_001136223
NM_001136224
NM_001136225
|
RCOR3
|
REST corepressor 3
|
|
chr20_-_3388219
|
0.195
|
NM_001009984
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr11_-_1771823
|
0.194
|
NM_001170820
|
IFITM10
|
interferon induced transmembrane protein 10
|
|
chr12_+_15699277
|
0.194
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr8_+_17354562
|
0.193
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr8_-_93111915
|
0.193
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr15_-_37393364
|
0.192
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr2_-_69614321
|
0.191
|
NM_001244710
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr5_+_76011780
|
0.189
|
NM_001992
|
F2R
|
coagulation factor II (thrombin) receptor
|
|
chr1_+_64239566
|
0.189
|
NM_001083592
NM_005012
|
ROR1
|
receptor tyrosine kinase-like orphan receptor 1
|
|
chr8_+_81398416
|
0.189
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr6_+_132129154
|
0.188
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr12_+_32654976
|
0.187
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr16_-_1429612
|
0.186
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr12_-_49177876
|
0.186
|
NM_015270
|
ADCY6
|
adenylate cyclase 6
|
|
chrX_-_77041678
|
0.185
|
NM_000489
NM_138270
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr16_+_2587951
|
0.185
|
NM_002613
NM_031268
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|