|
chr15_+_33010174
|
0.513
|
NM_001191322
NM_001191323
NM_013372
|
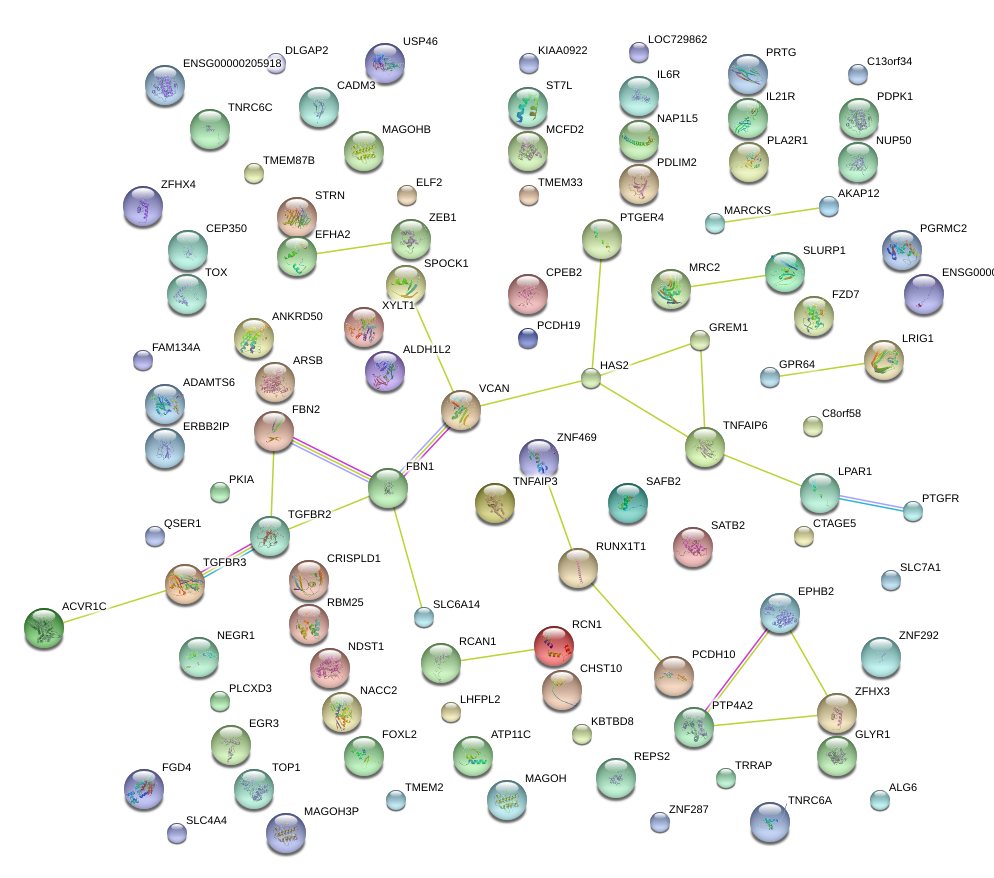
GREM1
|
gremlin 1
|
|
chr5_+_40680021
|
0.262
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr8_-_122653493
|
0.256
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr5_-_136834887
|
0.241
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr1_+_159141376
|
0.194
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr1_-_92351613
|
0.188
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr2_+_152214105
|
0.188
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr1_-_92371558
|
0.183
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr5_+_82767492
|
0.170
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr6_+_151646634
|
0.161
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr6_+_151561094
|
0.158
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr12_+_32654976
|
0.127
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr9_-_74383301
|
0.127
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr8_-_60031545
|
0.122
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr8_+_77593506
|
0.108
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr8_+_16884745
|
0.103
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr9_-_113800243
|
0.093
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr2_-_101034049
|
0.090
|
NM_004854
|
CHST10
|
carbohydrate sulfotransferase 10
|
|
chr16_+_27438578
|
0.088
|
NM_021798
|
IL21R
|
interleukin 21 receptor
|
|
chr1_-_113161724
|
0.085
|
NM_017744
NM_138727
NM_138728
NM_138729
|
ST7L
|
suppression of tumorigenicity 7 like
|
|
chr4_-_89618928
|
0.085
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr16_+_27413482
|
0.081
|
NM_181078
NM_181079
|
IL21R
|
interleukin 21 receptor
|
|
chr16_+_2587951
|
0.081
|
NM_002613
NM_031268
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr16_+_88493878
|
0.079
|
NM_001127464
|
ZNF469
|
zinc finger protein 469
|
|
chr1_+_63833260
|
0.078
|
NM_013339
|
ALG6
|
asparagine-linked glycosylation 6, alpha-1,3-glucosyltransferase homolog (S. cerevisiae)
|
|
chr2_-_200322699
|
0.077
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr15_-_48937795
|
0.073
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr4_+_15004297
|
0.073
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr2_-_200335988
|
0.071
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr6_+_138188352
|
0.069
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr5_+_149887673
|
0.069
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr1_+_179923870
|
0.068
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr1_+_23037329
|
0.068
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr5_-_78282356
|
0.065
|
NM_000046
|
ARSB
|
arylsulfatase B
|
|
chr20_+_39657461
|
0.065
|
NM_003286
|
TOP1
|
topoisomerase (DNA) I
|
|
chr2_-_200329810
|
0.064
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr16_-_17564737
|
0.063
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr8_+_22457098
|
0.063
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr7_+_98476084
|
0.062
|
NM_001244580
NM_003496
|
TRRAP
|
transformation/transcription domain-associated protein
|
|
chr6_+_114178516
|
0.061
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr8_-_93029864
|
0.060
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr13_-_30169795
|
0.059
|
NM_003045
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1
|
|
chr10_+_31608097
|
0.058
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr8_-_93115453
|
0.058
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_-_66550844
|
0.058
|
NM_015541
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr14_+_73525187
|
0.057
|
NM_021239
|
RBM25
|
RNA binding motif protein 25
|
|
chr4_+_72204769
|
0.055
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr4_-_129209983
|
0.054
|
NM_006320
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr10_+_31610063
|
0.053
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr22_+_45560529
|
0.053
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr4_+_154387450
|
0.053
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr8_-_93107442
|
0.052
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93111915
|
0.051
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_+_41937105
|
0.050
|
NM_018126
|
TMEM33
|
transmembrane protein 33
|
|
chrX_+_115567746
|
0.050
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr16_-_73082169
|
0.049
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr8_-_93107881
|
0.049
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_+_16964730
|
0.048
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr16_+_24740945
|
0.048
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr3_-_138665981
|
0.047
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr15_-_56035176
|
0.047
|
NM_173814
|
PRTG
|
protogenin
|
|
chr17_+_76000248
|
0.046
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr13_+_73301980
|
0.045
|
NM_024808
|
BORA
|
bora, aurora kinase A activator
|
|
chrX_-_19140581
|
0.045
|
NM_001079858
NM_001079859
NM_001079860
NM_001184833
NM_001184834
NM_001184835
NM_001184836
NM_001184837
NM_005756
|
GPR64
|
G protein-coupled receptor 64
|
|
chr2_+_202899309
|
0.044
|
NM_003507
|
FZD7
|
frizzled family receptor 7
|
|
chrX_-_99665270
|
0.043
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr2_+_220042901
|
0.043
|
NM_024293
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr14_+_39736277
|
0.043
|
NM_001247989
NM_001247990
NM_005930
NM_203355
|
CTAGE5
|
CTAGE family, member 5
|
|
chr8_+_79428335
|
0.043
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr9_-_138987096
|
0.043
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr11_+_32112414
|
0.042
|
NM_002901
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr2_-_160919120
|
0.041
|
NM_001007267
NM_001195641
NM_007366
|
PLA2R1
|
phospholipase A2 receptor 1, 180kDa
|
|
chr3_+_30647901
|
0.041
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr22_+_45559725
|
0.041
|
NM_007172
|
NUP50
|
nucleoporin 50kDa
|
|
chr11_+_32914359
|
0.040
|
NM_001076786
|
QSER1
|
glutamine and serine rich 1
|
|
chr2_-_47143250
|
0.040
|
NM_001171507
NM_001171510
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr1_+_154377668
|
0.040
|
NM_000565
NM_001206866
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr5_-_41510627
|
0.039
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr2_+_112812799
|
0.039
|
NM_032824
|
TMEM87B
|
transmembrane protein 87B
|
|
chr17_+_60704747
|
0.038
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr1_+_78956727
|
0.038
|
NM_000959
NM_001039585
|
PTGFR
|
prostaglandin F receptor (FP)
|
|
chr19_-_5622794
|
0.038
|
NM_014649
|
SAFB2
|
scaffold attachment factor B2
|
|
chr3_+_67048726
|
0.037
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr4_-_125633716
|
0.037
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr5_-_64777703
|
0.037
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr2_-_47142950
|
0.035
|
NM_001171506
NM_001171509
NM_139279
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr8_-_93075190
|
0.035
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_72748147
|
0.035
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr1_-_32385258
|
0.035
|
NM_001195100
NM_001195101
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr1_-_32403975
|
0.034
|
NM_080391
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr12_-_105478218
|
0.034
|
NM_001034173
|
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2
|
|
chrX_-_138914384
|
0.033
|
NM_001010986
NM_173694
|
ATP11C
|
ATPase, class VI, type 11C
|
|
chr17_-_16472463
|
0.032
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chr8_+_1449531
|
0.032
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr5_+_65222381
|
0.032
|
NM_001006600
NM_001253697
NM_001253698
NM_001253699
NM_001253701
NM_018695
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr2_-_37193609
|
0.032
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr16_-_4897265
|
0.032
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr4_-_53522756
|
0.031
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr8_-_22550057
|
0.031
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr5_-_127873734
|
0.030
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr4_-_140005464
|
0.030
|
NM_006874
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr5_-_77944341
|
0.030
|
NM_005779
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr8_-_22549901
|
0.030
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr2_-_158485393
|
0.029
|
NM_001111032
NM_001111033
NM_145259
|
ACVR1C
|
activin A receptor, type IC
|
|
chr12_-_10766130
|
0.029
|
NM_018048
|
MAGOHB
|
mago-nashi homolog B (Drosophila)
|
|
chr2_-_47168902
|
0.029
|
NM_001171508
NM_001171511
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr6_+_87865261
|
0.029
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr8_+_75896707
|
0.029
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr13_-_80915041
|
0.029
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr6_-_24719286
|
0.028
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr16_-_73092533
|
0.028
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr8_-_22550708
|
0.028
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr14_-_61115906
|
0.028
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr10_-_65028825
|
0.027
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr11_+_133938819
|
0.027
|
NM_001205329
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr15_-_56209305
|
0.027
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr5_+_173315313
|
0.027
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr11_+_69455837
|
0.027
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr4_+_140222668
|
0.026
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr17_+_75084724
|
0.026
|
NM_001204408
|
SEC14L1
LINC00338
|
SEC14-like 1 (S. cerevisiae)
long intergenic non-protein coding RNA 338
|
|
chr12_-_80307690
|
0.026
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr16_-_52580805
|
0.026
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr2_-_1748285
|
0.026
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr14_+_39734475
|
0.026
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr2_-_158454025
|
0.025
|
NM_001111031
|
ACVR1C
|
activin A receptor, type IC
|
|
chr7_-_97881436
|
0.025
|
NM_015395
|
TECPR1
|
tectonin beta-propeller repeat containing 1
|
|
chr15_-_56285732
|
0.025
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr20_+_5107393
|
0.025
|
NM_003818
|
CDS2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2
|
|
chr14_-_74485813
|
0.025
|
NM_001249
|
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5
|
|
chrX_+_113818550
|
0.024
|
NM_000868
|
HTR2C
|
5-hydroxytryptamine (serotonin) receptor 2C
|
|
chrX_-_83757398
|
0.024
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr5_-_157002739
|
0.024
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr12_+_122242580
|
0.024
|
NM_015048
|
SETD1B
|
SET domain containing 1B
|
|
chr3_+_77089248
|
0.023
|
NM_002942
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila)
|
|
chr16_-_52581713
|
0.023
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr4_-_53525316
|
0.023
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr12_-_9102262
|
0.023
|
NM_001207024
NM_002355
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr11_-_85376159
|
0.023
|
NM_001039618
|
CREBZF
|
CREB/ATF bZIP transcription factor
|
|
chr1_+_90098643
|
0.023
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr12_+_22778008
|
0.023
|
NM_001039481
NM_018638
|
ETNK1
|
ethanolamine kinase 1
|
|
chr4_+_160188997
|
0.023
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr3_+_140770742
|
0.023
|
NM_080862
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4
|
|
chr2_-_179343225
|
0.022
|
NM_001135212
NM_181342
|
FKBP7
|
FK506 binding protein 7
|
|
chr1_+_65730423
|
0.022
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr12_-_46663207
|
0.022
|
NM_001077484
NM_030674
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr22_+_40440664
|
0.022
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr1_+_198125890
|
0.022
|
NM_133494
|
NEK7
|
NIMA (never in mitosis gene a)-related kinase 7
|
|
chr1_-_149889362
|
0.022
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr1_-_225615787
|
0.021
|
NM_002296
|
LBR
|
lamin B receptor
|
|
chrX_-_154493736
|
0.021
|
NM_171998
|
RAB39B
|
RAB39B, member RAS oncogene family
|
|
chr5_+_127419407
|
0.021
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr5_-_79551832
|
0.021
|
NM_001174071
NM_001174072
NM_178276
|
SERINC5
|
serine incorporator 5
|
|
chr22_+_29313376
|
0.021
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr17_-_27621163
|
0.021
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr12_-_42538432
|
0.020
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr7_-_14029641
|
0.020
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr17_-_66287256
|
0.020
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr6_-_116575170
|
0.020
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr11_-_66115016
|
0.020
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr17_+_75085234
|
0.020
|
NM_001204410
|
SEC14L1
LINC00338
|
SEC14-like 1 (S. cerevisiae)
long intergenic non-protein coding RNA 338
|
|
chr7_-_25164902
|
0.020
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr14_+_52327021
|
0.020
|
NM_001243773
NM_001243774
NM_053064
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr12_-_80329234
|
0.020
|
NM_001143885
NM_001244990
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr17_+_47074749
|
0.019
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr10_+_124768401
|
0.019
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr3_-_52713728
|
0.019
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr12_-_104234968
|
0.019
|
NM_001031701
|
NT5DC3
|
5'-nucleotidase domain containing 3
|
|
chr20_+_43595119
|
0.019
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr7_+_64254763
|
0.019
|
NM_001160183
NM_006524
|
ZNF138
|
zinc finger protein 138
|
|
chr1_-_225616556
|
0.019
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr11_+_86748870
|
0.019
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr2_+_24807345
|
0.019
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr22_+_29279573
|
0.019
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr7_-_14030864
|
0.019
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr5_-_55290601
|
0.018
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr17_+_45608443
|
0.018
|
NM_006310
|
NPEPPS
|
aminopeptidase puromycin sensitive
|
|
chr9_-_95432542
|
0.018
|
NM_022755
|
IPPK
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase
|
|
chr2_+_85766100
|
0.018
|
NM_005911
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr20_+_43990576
|
0.018
|
NM_001197129
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr7_-_14031049
|
0.018
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr3_-_52719514
|
0.018
|
NM_018313
|
PBRM1
|
polybromo 1
|
|
chr11_+_94277005
|
0.018
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr2_+_172950207
|
0.018
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr10_-_65225605
|
0.018
|
NM_032776
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr18_+_67956136
|
0.018
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr2_-_10952855
|
0.017
|
NM_005742
|
PDIA6
|
protein disulfide isomerase family A, member 6
|
|
chrX_+_9754465
|
0.017
|
NM_001649
|
SHROOM2
|
shroom family member 2
|
|
chr11_-_96122984
|
0.017
|
NM_024725
|
CCDC82
|
coiled-coil domain containing 82
|
|
chr7_-_14025826
|
0.017
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr20_+_31407696
|
0.017
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr3_-_69062744
|
0.017
|
NM_173654
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr3_+_119187782
|
0.017
|
NM_152305
|
POGLUT1
|
protein O-glucosyltransferase 1
|
|
chr1_-_212004113
|
0.017
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr10_+_1102741
|
0.017
|
NM_014023
|
WDR37
|
WD repeat domain 37
|
|
chr4_+_72052354
|
0.016
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr1_-_46598250
|
0.016
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr6_+_10555948
|
0.016
|
NM_001491
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr5_-_131132715
|
0.016
|
NM_001008738
NM_133372
|
FNIP1
|
folliculin interacting protein 1
|
|
chr2_+_207139413
|
0.016
|
NM_020923
|
ZDBF2
|
zinc finger, DBF-type containing 2
|
|
chr2_-_70475563
|
0.016
|
NM_022037
NM_022173
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|