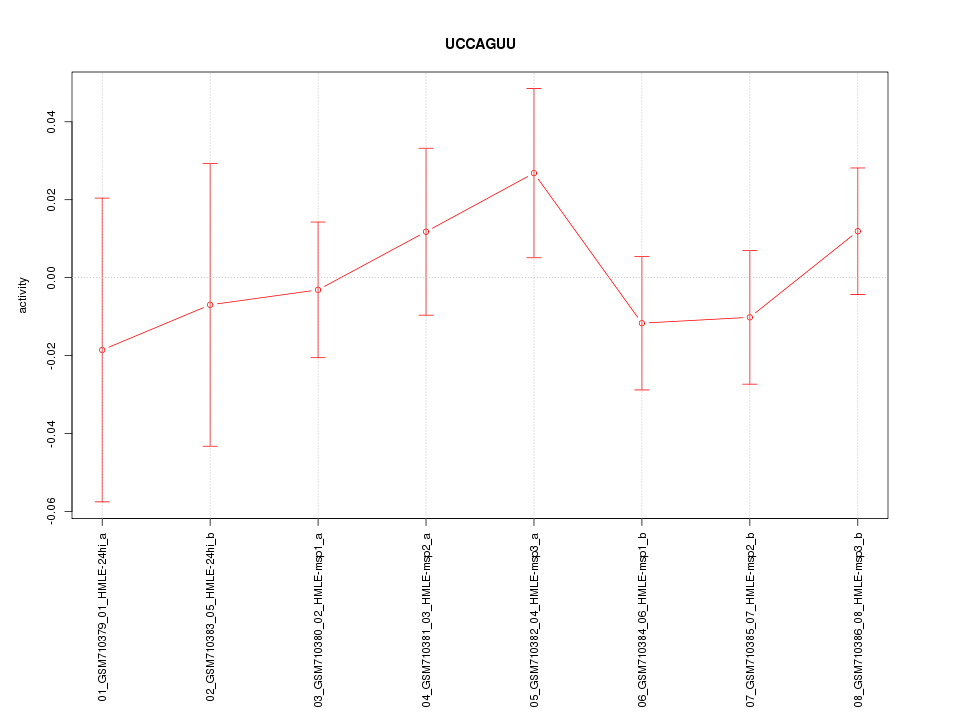
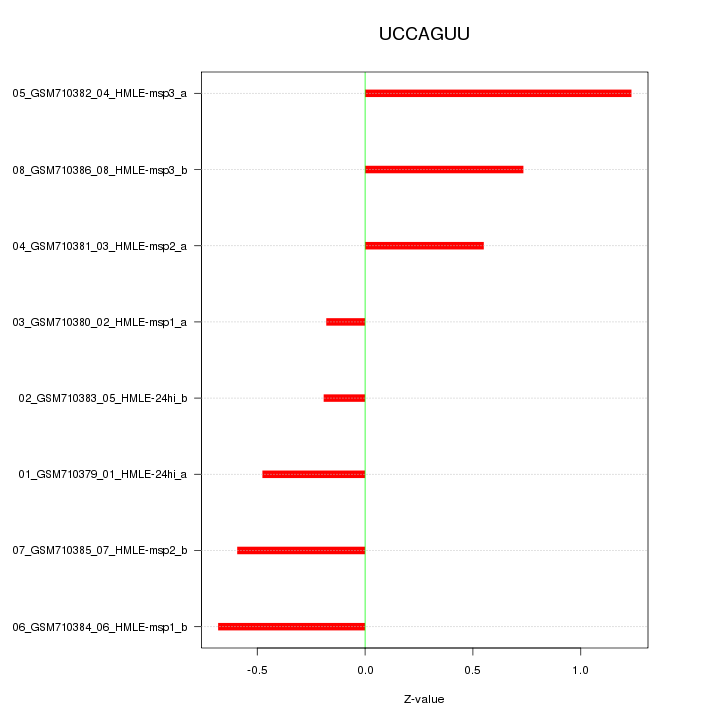
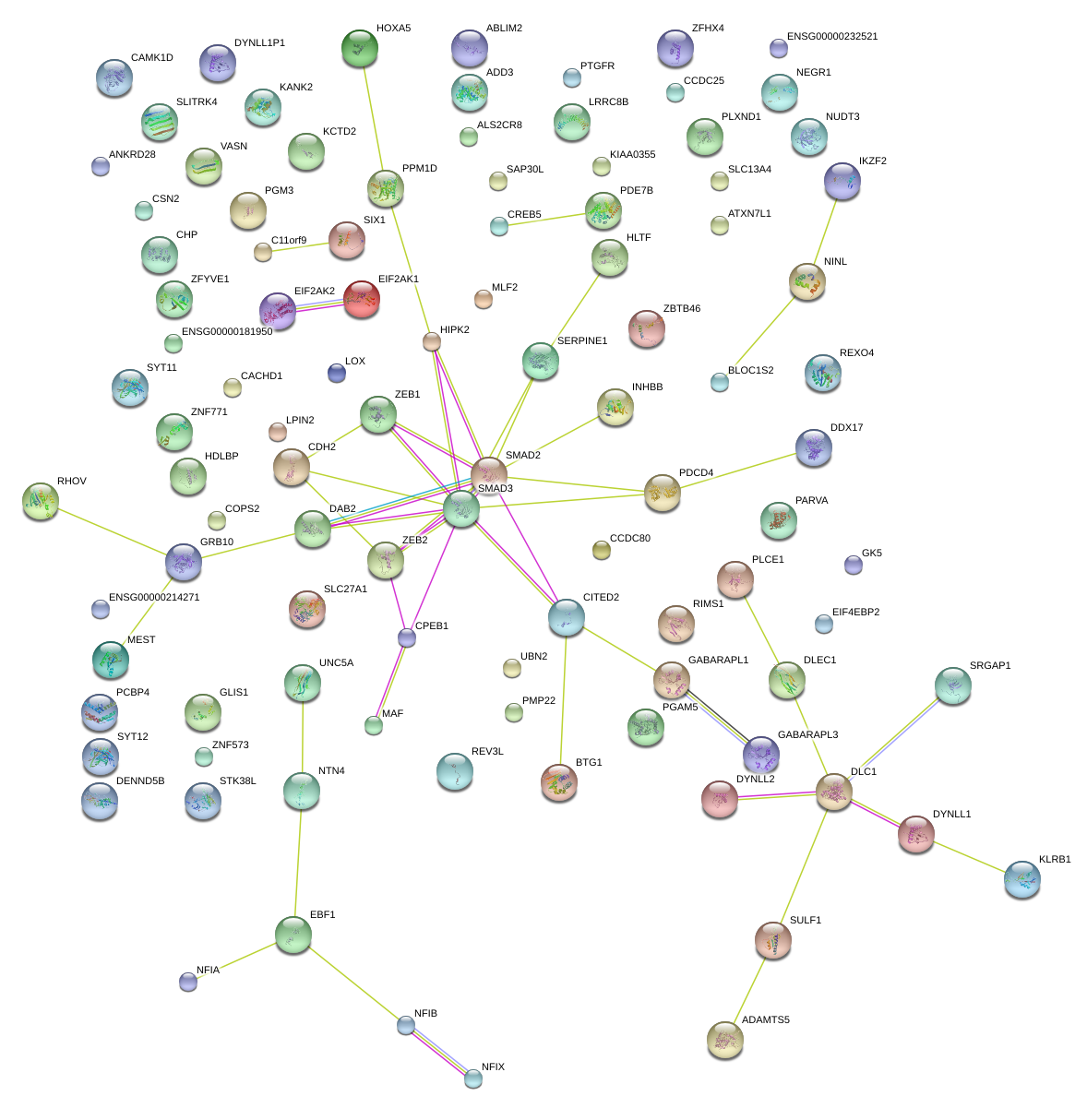
|
chr1_+_78956727
|
0.866
|
NM_000959
NM_001039585
|
PTGFR
|
prostaglandin F receptor (FP)
|
|
chr6_+_72926462
|
0.825
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_+_72922513
|
0.806
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_+_72926144
|
0.782
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_+_73076431
|
0.752
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_89990396
|
0.451
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr1_-_72748147
|
0.421
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr10_+_95848811
|
0.416
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr10_+_95753684
|
0.413
|
NM_016341
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr1_-_54199876
|
0.410
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr1_+_90024271
|
0.391
|
NM_015350
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr8_-_12973752
|
0.389
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr5_-_158526698
|
0.385
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr6_+_72596648
|
0.370
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr8_-_12990731
|
0.359
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr2_-_214016332
|
0.327
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chrX_-_142722869
|
0.313
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chrX_-_142722318
|
0.305
|
NM_001184750
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr8_+_70405027
|
0.297
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr7_-_50861114
|
0.282
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chrX_-_142723921
|
0.275
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr10_+_111767685
|
0.274
|
NM_001121
NM_016824
|
ADD3
|
adducin 3 (gamma)
|
|
chr8_+_70378858
|
0.264
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr5_-_39425297
|
0.263
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr3_-_129325175
|
0.251
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr12_-_96184510
|
0.248
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr7_-_50800018
|
0.246
|
NM_001001549
NM_005311
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr14_-_73453651
|
0.235
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr6_+_136172802
|
0.230
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr6_-_139695349
|
0.230
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr8_+_77593506
|
0.229
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr2_-_145277879
|
0.228
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr6_-_139695784
|
0.226
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr12_-_92539614
|
0.226
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr7_-_50772997
|
0.225
|
NM_001001550
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr5_-_121412805
|
0.218
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr14_-_73493744
|
0.218
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr7_-_139477437
|
0.217
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr5_-_121414011
|
0.216
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr18_-_25757351
|
0.214
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr4_-_8160435
|
0.207
|
NM_001130083
NM_001130084
NM_001130085
NM_001130086
NM_001130087
NM_001130088
NM_032432
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr20_-_25566137
|
0.205
|
NM_025176
|
NINL
|
ninein-like
|
|
chr7_-_27183225
|
0.204
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr5_+_153825497
|
0.202
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr10_+_111765725
|
0.193
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr19_+_34745452
|
0.189
|
NM_014686
|
KIAA0355
|
KIAA0355
|
|
chr15_-_52970808
|
0.185
|
NM_019600
|
FAM214A
|
family with sequence similarity 214, member A
|
|
chr18_-_3011944
|
0.180
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr20_-_62436855
|
0.175
|
NM_025224
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr2_-_214015053
|
0.170
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr11_+_66790189
|
0.165
|
NM_001177880
|
SYT12
|
synaptotagmin XII
|
|
chr7_+_130126035
|
0.164
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr21_-_28339405
|
0.163
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr10_+_12391471
|
0.154
|
NM_020397
NM_153498
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr8_-_27630096
|
0.153
|
NM_018246
|
CCDC25
|
coiled-coil domain containing 25
|
|
chr12_-_31743822
|
0.152
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr3_-_52001388
|
0.152
|
NM_020418
NM_001174100
NM_033008
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr3_-_15900928
|
0.150
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr3_-_51996855
|
0.149
|
NM_033010
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr12_+_10365415
|
0.148
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr11_+_66790815
|
0.148
|
NM_177963
|
SYT12
|
synaptotagmin XII
|
|
chr7_+_130131921
|
0.147
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr7_+_130131172
|
0.142
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr16_+_4421774
|
0.141
|
NM_138440
|
VASN
|
vasorin
|
|
chr3_-_141944441
|
0.141
|
NM_001039547
|
GK5
|
glycerol kinase 5 (putative)
|
|
chr12_+_27397077
|
0.137
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr19_-_11308184
|
0.134
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr11_+_61522860
|
0.134
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr7_-_105319608
|
0.133
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr7_+_28475233
|
0.132
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr11_+_61520000
|
0.132
|
NM_001127392
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr2_+_121103670
|
0.131
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr15_+_41523401
|
0.130
|
NM_007236
|
CHP
|
calcium binding protein P22
|
|
chr17_+_58677489
|
0.127
|
NM_003620
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr3_-_112359962
|
0.120
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_+_28338939
|
0.115
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr10_-_102045948
|
0.115
|
NM_001001342
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2
|
|
chr11_+_12399024
|
0.114
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr6_-_83903632
|
0.113
|
NM_001199917
|
PGM3
|
phosphoglucomutase 3
|
|
chr17_-_15164043
|
0.112
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr6_-_111804413
|
0.112
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr17_-_15165873
|
0.110
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr15_-_83316621
|
0.109
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr14_-_61115906
|
0.108
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr7_+_100770378
|
0.108
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr17_-_15168590
|
0.108
|
NM_000304
|
PMP22
|
peripheral myelin protein 22
|
|
chr10_-_102046415
|
0.107
|
NM_173809
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2
|
|
chr19_-_11305186
|
0.106
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr10_+_72163860
|
0.106
|
NM_004096
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr7_+_28452143
|
0.103
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr15_+_67358194
|
0.102
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr16_-_79634610
|
0.102
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr7_+_138916230
|
0.100
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr7_+_28725597
|
0.100
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr3_-_148804122
|
0.098
|
NM_003071
NM_139048
|
HLTF
|
helicase-like transcription factor
|
|
chr2_+_203776930
|
0.098
|
NM_001104586
NM_024744
|
ALS2CR8
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 8
|
|
chr10_+_112631552
|
0.098
|
NM_001199492
NM_014456
NM_145341
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr12_+_64238511
|
0.097
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr6_-_34360438
|
0.097
|
NM_006703
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3
|
|
chr7_-_6098813
|
0.097
|
NM_001134335
NM_014413
|
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1
|
|
chr2_-_242255114
|
0.096
|
NM_005336
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr22_-_38902344
|
0.095
|
NM_001098504
NM_006386
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr4_-_70826712
|
0.094
|
NM_001891
|
CSN2
|
casein beta
|
|
chr1_+_64936475
|
0.093
|
NM_020925
|
CACHD1
|
cache domain containing 1
|
|
chr1_+_61547625
|
0.092
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr17_+_73043154
|
0.091
|
NM_015353
|
KCTD2
|
potassium channel tetramerisation domain containing 2
|
|
chr7_+_135347211
|
0.090
|
NM_001130929
|
SLC13A4
C7orf73
|
solute carrier family 13 (sodium/sulfate symporters), member 4
chromosome 7 open reading frame 73
|
|
chr17_+_33914278
|
0.089
|
NM_001030006
NM_001282
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr8_-_13372183
|
0.089
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr5_-_180688095
|
0.088
|
NM_032765
|
TRIM52
|
tripartite motif containing 52
|
|
chr13_+_103451398
|
0.088
|
NM_001159596
NM_017693
|
BIVM
|
basic, immunoglobulin-like variable motif containing
|
|
chrX_-_83442906
|
0.085
|
NM_014496
|
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6
|
|
chr12_+_72233486
|
0.085
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr2_-_219432955
|
0.085
|
NM_020935
|
USP37
|
ubiquitin specific peptidase 37
|
|
chr8_-_121824287
|
0.084
|
NM_021021
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr21_+_45285101
|
0.084
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr12_+_93771607
|
0.083
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr5_+_122181159
|
0.082
|
NM_014035
|
SNX24
|
sorting nexin 24
|
|
chr3_+_16926451
|
0.082
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr21_+_45345278
|
0.081
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr12_+_131438451
|
0.081
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr19_+_18794391
|
0.080
|
NM_001098482
NM_015321
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr15_+_62939509
|
0.080
|
NM_015059
|
TLN2
|
talin 2
|
|
chr16_-_49860917
|
0.078
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr1_-_21605984
|
0.077
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr17_+_21279667
|
0.076
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr17_-_58603492
|
0.076
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr11_+_64794879
|
0.076
|
NM_013306
NM_147777
|
SNX15
|
sorting nexin 15
|
|
chr9_-_95896498
|
0.076
|
NM_004148
|
NINJ1
|
ninjurin 1
|
|
chr17_+_7452374
|
0.075
|
NM_172089
NM_003809
|
TNFSF12-TNFSF13
TNFSF12
|
TNFSF12-TNFSF13 readthrough
tumor necrosis factor (ligand) superfamily, member 12
|
|
chr1_-_21616648
|
0.074
|
NM_001113349
|
ECE1
|
endothelin converting enzyme 1
|
|
chr13_+_24144402
|
0.073
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr2_-_227663454
|
0.073
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr9_+_120466453
|
0.073
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr17_+_61699786
|
0.072
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr14_-_45431121
|
0.072
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr4_-_114682836
|
0.072
|
NM_001221
NM_172114
NM_172115
NM_172127
NM_172128
NM_172129
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr1_-_183604885
|
0.072
|
NM_005717
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr8_-_74791098
|
0.071
|
NM_001001481
NM_018299
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr14_-_35099296
|
0.071
|
NM_021249
NM_152233
|
SNX6
|
sorting nexin 6
|
|
chr4_+_86396251
|
0.071
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr10_-_118032717
|
0.070
|
NM_001145453
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr4_+_160188997
|
0.070
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr4_-_78740508
|
0.070
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr14_-_35183722
|
0.070
|
NM_001243645
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr12_-_76478737
|
0.069
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr8_+_26435339
|
0.069
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr19_+_1269244
|
0.068
|
NM_001280
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr4_+_154074269
|
0.068
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr2_-_27718098
|
0.067
|
NM_022823
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr15_-_83240499
|
0.067
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr2_-_242212244
|
0.067
|
NM_001243900
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr14_-_54955714
|
0.067
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr1_-_159895279
|
0.067
|
NM_003564
|
TAGLN2
|
transgelin 2
|
|
chr5_-_171710794
|
0.066
|
NM_152277
|
UBTD2
|
ubiquitin domain containing 2
|
|
chr18_-_70534809
|
0.066
|
NM_001201465
NM_138966
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr17_-_1012257
|
0.066
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr7_+_117120016
|
0.066
|
NM_000492
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7)
|
|
chr11_-_64570621
|
0.066
|
NM_004579
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2
|
|
chr16_+_69220940
|
0.066
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr3_-_15839674
|
0.066
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr9_-_20622476
|
0.065
|
NM_004529
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr12_-_110505499
|
0.065
|
NM_207435
|
C12orf76
|
chromosome 12 open reading frame 76
|
|
chr12_-_56727720
|
0.065
|
NM_001166279
NM_014871
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr4_-_140060601
|
0.065
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr4_+_86851425
|
0.064
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr16_+_75600248
|
0.064
|
NM_007285
|
GABARAPL2
|
GABA(A) receptor-associated protein-like 2
|
|
chr17_-_1090562
|
0.064
|
NM_001159746
|
ABR
|
active BCR-related gene
|
|
chr13_-_41240607
|
0.062
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr14_-_35182944
|
0.062
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr20_-_33460558
|
0.062
|
NM_178026
|
GGT7
|
gamma-glutamyltransferase 7
|
|
chr5_+_60628085
|
0.061
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr4_+_154125589
|
0.060
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr6_+_96463844
|
0.059
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr9_+_4679557
|
0.059
|
NM_017913
|
CDC37L1
|
cell division cycle 37 homolog (S. cerevisiae)-like 1
|
|
chr15_+_67458492
|
0.059
|
NM_001145104
|
SMAD3
|
SMAD family member 3
|
|
chr8_+_26371461
|
0.059
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr1_-_21671905
|
0.059
|
NM_001113348
|
ECE1
|
endothelin converting enzyme 1
|
|
chr1_+_61542928
|
0.059
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr7_-_105516923
|
0.058
|
NM_020725
NM_152749
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr2_-_128100673
|
0.058
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr12_+_49760679
|
0.057
|
NM_023071
|
SPATS2
|
spermatogenesis associated, serine-rich 2
|
|
chr21_+_38792600
|
0.057
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr19_-_58662144
|
0.057
|
NM_024620
|
ZNF329
|
zinc finger protein 329
|
|
chr10_+_94449678
|
0.057
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr12_-_56727446
|
0.056
|
NM_001127460
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr5_+_135468522
|
0.056
|
NM_001001419
NM_001001420
NM_005903
|
SMAD5
|
SMAD family member 5
|
|
chr11_-_9025531
|
0.055
|
NM_020645
|
NRIP3
|
nuclear receptor interacting protein 3
|
|
chr1_+_162466963
|
0.054
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr1_-_171711093
|
0.054
|
NM_001185127
NM_003762
|
VAMP4
|
vesicle-associated membrane protein 4
|
|
chr10_-_118032975
|
0.053
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr5_-_179233772
|
0.053
|
NM_014275
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr11_-_104035023
|
0.053
|
NM_025208
NM_033135
|
PDGFD
|
platelet derived growth factor D
|
|
chr13_+_49549953
|
0.053
|
NM_001079673
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr17_+_7608413
|
0.053
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr21_-_34100228
|
0.053
|
NM_001160302
NM_001160306
|
SYNJ1
|
synaptojanin 1
|
|
chr12_+_53845884
|
0.052
|
NM_001098620
NM_001128911
NM_001128912
NM_001128913
NM_001128914
NM_005016
NM_031989
|
PCBP2
|
poly(rC) binding protein 2
|
|
chr1_-_32403975
|
0.051
|
NM_080391
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr19_-_11670003
|
0.051
|
NM_032377
|
ELOF1
|
elongation factor 1 homolog (S. cerevisiae)
|
|
chr2_-_204399906
|
0.051
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|