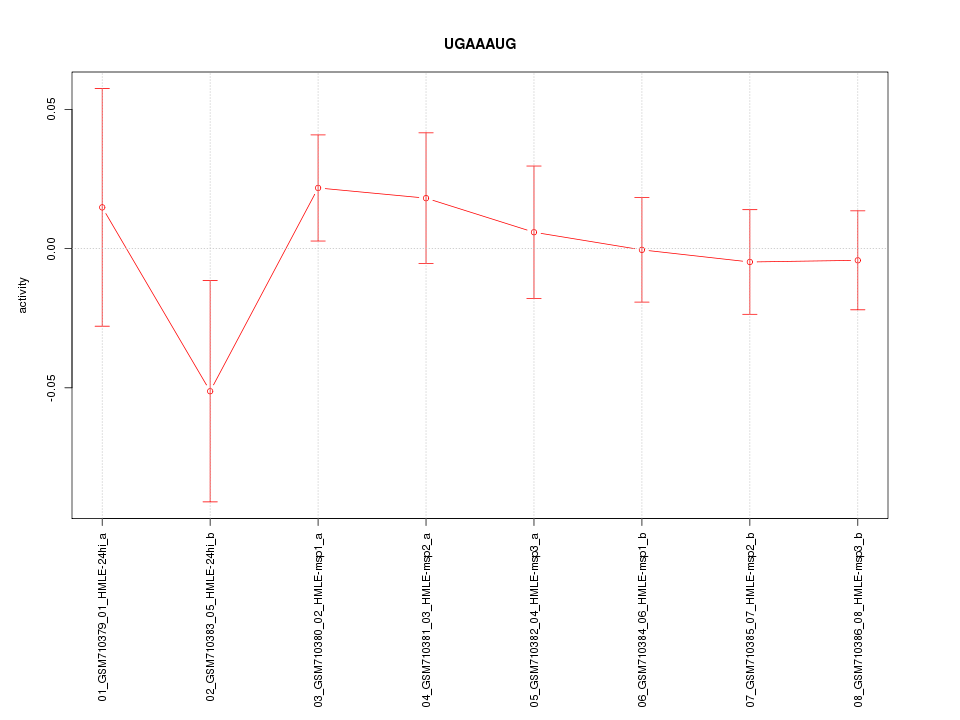
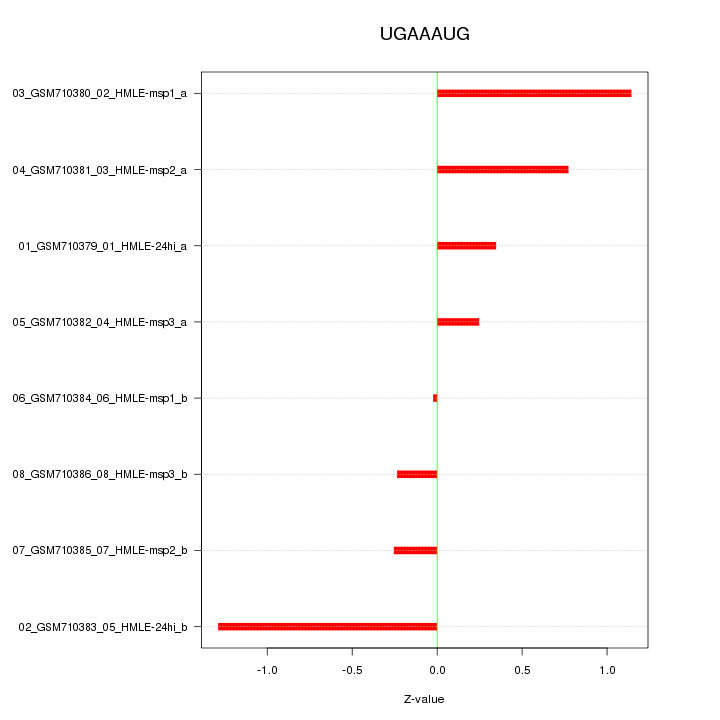
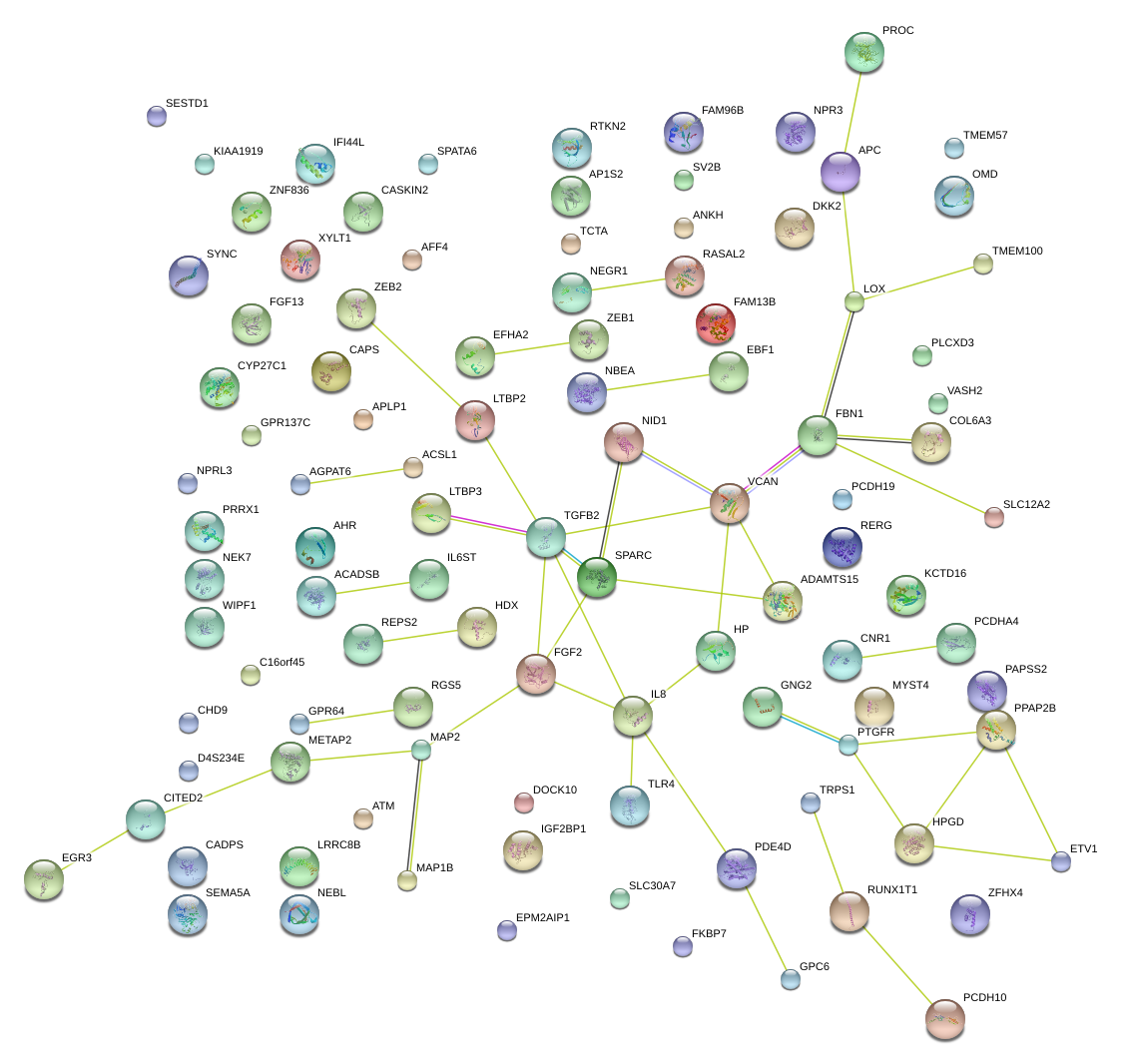
|
chr5_-_9546157
|
0.500
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr10_+_31608097
|
0.466
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr10_+_31610063
|
0.433
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr10_-_21463019
|
0.383
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr1_-_72748147
|
0.381
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr1_-_57045235
|
0.367
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr1_-_236228476
|
0.365
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr5_+_32711437
|
0.339
|
NM_000908
NM_001204375
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr15_-_48937795
|
0.327
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr5_+_32710742
|
0.326
|
NM_001204376
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr10_-_21186530
|
0.274
|
NM_006393
|
NEBL
|
nebulette
|
|
chr5_+_82767492
|
0.272
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr5_-_137368778
|
0.265
|
NM_001101800
NM_001101801
NM_016603
|
FAM13B
|
family with sequence similarity 13, member B
|
|
chr1_+_178310605
|
0.259
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chrX_-_99665270
|
0.244
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr5_-_158526698
|
0.235
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr4_-_185747183
|
0.232
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr5_-_151066492
|
0.232
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr8_+_41435666
|
0.228
|
NM_178819
|
AGPAT6
|
1-acylglycerol-3-phosphate O-acyltransferase 6 (lysophosphatidic acid acyltransferase, zeta)
|
|
chr9_+_120466453
|
0.222
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr1_+_178062829
|
0.217
|
NM_170692
|
RASAL2
|
RAS protein activator like 2
|
|
chr13_+_93879011
|
0.215
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr1_+_78956727
|
0.204
|
NM_000959
NM_001039585
|
PTGFR
|
prostaglandin F receptor (FP)
|
|
chr14_+_52327021
|
0.198
|
NM_001243773
NM_001243774
NM_053064
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr5_+_143550395
|
0.192
|
NM_020768
|
KCTD16
|
potassium channel tetramerisation domain containing 16
|
|
chr12_-_15374290
|
0.192
|
NM_001190726
NM_032918
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor
|
|
chr6_-_139695349
|
0.192
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr4_+_4387982
|
0.190
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr5_+_112043188
|
0.188
|
NM_001127511
|
APC
|
adenomatous polyposis coli
|
|
chr6_-_139695784
|
0.188
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr2_-_180129245
|
0.186
|
NM_178123
|
SESTD1
|
SEC14 and spectrin domains 1
|
|
chr8_+_16884745
|
0.181
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr6_-_88875766
|
0.181
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr1_-_163172624
|
0.179
|
NM_001195303
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr8_-_22549901
|
0.178
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_22550708
|
0.177
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr8_-_22550057
|
0.175
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr4_-_175443791
|
0.174
|
NM_000860
NM_001145816
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD)
|
|
chr1_-_163172863
|
0.172
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr5_+_112073555
|
0.171
|
NM_000038
NM_001127510
|
APC
|
adenomatous polyposis coli
|
|
chr1_+_170633312
|
0.169
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr5_-_132299263
|
0.169
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr1_-_33168356
|
0.168
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr2_-_145277879
|
0.163
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_179343225
|
0.158
|
NM_001135212
NM_181342
|
FKBP7
|
FK506 binding protein 7
|
|
chr17_+_47074749
|
0.156
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr5_+_71403040
|
0.156
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr5_-_58334936
|
0.153
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_55290601
|
0.152
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr4_+_123747815
|
0.149
|
NM_002006
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr5_-_121412805
|
0.149
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr14_-_75078669
|
0.148
|
NM_000428
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr11_+_108093558
|
0.147
|
NM_000051
|
ATM
|
ataxia telangiectasia mutated
|
|
chr10_-_64028457
|
0.146
|
NM_145307
|
RTKN2
|
rhotekin 2
|
|
chr5_-_121414011
|
0.144
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr1_+_213123853
|
0.143
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chrX_+_16964730
|
0.141
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr2_-_127963342
|
0.137
|
NM_001001665
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1
|
|
chr15_+_91643429
|
0.135
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr2_-_175547471
|
0.134
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr16_+_15596122
|
0.134
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr10_+_89419352
|
0.133
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chrX_-_19140581
|
0.132
|
NM_001079858
NM_001079859
NM_001079860
NM_001184833
NM_001184834
NM_001184835
NM_001184836
NM_001184837
NM_005756
|
GPR64
|
G protein-coupled receptor 64
|
|
chr16_-_17564737
|
0.132
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr8_-_93115453
|
0.132
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_-_83757398
|
0.131
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr7_+_17338182
|
0.130
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr2_-_175499275
|
0.129
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr16_+_15528324
|
0.127
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr8_+_77593506
|
0.127
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr1_-_48937791
|
0.125
|
NM_019073
|
SPATA6
|
spermatogenesis associated 6
|
|
chr11_+_130318457
|
0.124
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr1_+_89990396
|
0.124
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr1_+_79086087
|
0.124
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr5_+_127419407
|
0.123
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr1_+_198125890
|
0.123
|
NM_133494
|
NEK7
|
NIMA (never in mitosis gene a)-related kinase 7
|
|
chr3_-_37034609
|
0.122
|
NM_014805
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1
|
|
chr17_-_53809444
|
0.122
|
NM_001099640
|
TMEM100
|
transmembrane protein 100
|
|
chr5_-_14871555
|
0.121
|
NM_054027
|
ANKH
|
ankylosis, progressive homolog (mouse)
|
|
chr17_-_53800074
|
0.120
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chrX_-_15873094
|
0.120
|
NM_003916
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr4_+_74606222
|
0.120
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr13_+_35516390
|
0.119
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr14_+_53019865
|
0.118
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr10_+_76586299
|
0.118
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr19_+_36359346
|
0.117
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr1_+_101361589
|
0.116
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr5_-_41510627
|
0.115
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr1_+_25757381
|
0.115
|
NM_018202
|
TMEM57
|
transmembrane protein 57
|
|
chr7_-_14031049
|
0.115
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr8_-_93029864
|
0.115
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_225907312
|
0.114
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr4_-_107957452
|
0.114
|
NM_014421
|
DKK2
|
dickkopf 2 homolog (Xenopus laevis)
|
|
chr16_+_53088791
|
0.113
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr9_-_95186559
|
0.112
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr8_-_116681103
|
0.112
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr19_+_5914192
|
0.112
|
NM_004058
NM_080590
|
CAPS
|
calcyphosine
|
|
chr8_-_93107442
|
0.112
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_+_210288668
|
0.112
|
NM_001039538
|
MAP2
|
microtubule-associated protein 2
|
|
chr1_+_218518675
|
0.112
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr8_-_93107881
|
0.110
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr6_+_111580481
|
0.110
|
NM_153369
|
KIAA1919
|
KIAA1919
|
|
chr3_+_49449638
|
0.110
|
NM_022171
|
TCTA
|
T-cell leukemia translocation altered gene
|
|
chr10_+_124768401
|
0.109
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr2_-_238322840
|
0.109
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr3_+_185080835
|
0.109
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr8_-_93111915
|
0.108
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_90024271
|
0.107
|
NM_015350
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr2_+_210444364
|
0.106
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chrX_+_7137471
|
0.104
|
NM_000351
|
STS
|
steroid sulfatase (microsomal), isozyme S
|
|
chr5_+_49962771
|
0.103
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr4_+_157997181
|
0.103
|
NM_000824
NM_001166061
NM_001166060
|
GLRB
|
glycine receptor, beta
|
|
chr2_+_172378856
|
0.103
|
NM_001127383
NM_024843
|
CYBRD1
|
cytochrome b reductase 1
|
|
chr1_-_183387501
|
0.102
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr14_+_85996454
|
0.102
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr20_+_5107393
|
0.102
|
NM_003818
|
CDS2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2
|
|
chr8_+_70405027
|
0.101
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr5_+_153570270
|
0.101
|
NM_198321
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10)
|
|
chrX_-_154114431
|
0.100
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chrX_+_113818550
|
0.100
|
NM_000868
|
HTR2C
|
5-hydroxytryptamine (serotonin) receptor 2C
|
|
chr5_-_138211054
|
0.099
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr4_-_96470350
|
0.099
|
NM_003728
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr7_+_8008416
|
0.099
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr13_-_49107250
|
0.098
|
NM_001268
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2
|
|
chr12_+_75874512
|
0.098
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr9_-_79520878
|
0.098
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr7_+_7222142
|
0.098
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr7_-_14029641
|
0.097
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr1_-_183274008
|
0.096
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr8_+_97505876
|
0.096
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr9_+_118916069
|
0.096
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr15_+_63569748
|
0.096
|
NM_001145646
NM_031301
|
APH1B
|
anterior pharynx defective 1 homolog B (C. elegans)
|
|
chr8_+_95732101
|
0.096
|
NM_181787
|
DPY19L4
|
dpy-19-like 4 (C. elegans)
|
|
chr2_+_109745996
|
0.095
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr21_-_39288661
|
0.094
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr8_-_139509064
|
0.094
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chrX_-_154493736
|
0.094
|
NM_171998
|
RAB39B
|
RAB39B, member RAS oncogene family
|
|
chr7_+_70597788
|
0.094
|
NM_022479
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17
|
|
chr13_-_33760215
|
0.094
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr7_-_14030864
|
0.093
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr12_+_65672682
|
0.092
|
NM_001193461
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr12_-_92539614
|
0.092
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr5_-_58295758
|
0.092
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_149340287
|
0.092
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr5_-_150537335
|
0.092
|
NM_001155
|
ANXA6
|
annexin A6
|
|
chr17_-_76356153
|
0.091
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chrX_+_103031753
|
0.091
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr13_-_33859835
|
0.091
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr13_-_33780142
|
0.090
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr9_+_35732207
|
0.090
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr6_+_24495166
|
0.090
|
NM_001080
NM_170740
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1
|
|
chrX_-_154250769
|
0.089
|
NM_000132
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chrX_+_103031433
|
0.089
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr9_-_98269686
|
0.089
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr11_-_45307617
|
0.089
|
NM_001247987
NM_020826
|
SYT13
|
synaptotagmin XIII
|
|
chr3_+_169684549
|
0.089
|
NM_003262
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr7_-_131241321
|
0.088
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr5_-_150521200
|
0.088
|
NM_001193544
|
ANXA6
|
annexin A6
|
|
chr2_-_55459406
|
0.087
|
NM_152385
|
C2orf63
|
chromosome 2 open reading frame 63
|
|
chr9_-_123342258
|
0.087
|
NM_001011649
NM_018249
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2
|
|
chr2_-_227663454
|
0.087
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr5_-_95768660
|
0.086
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr22_-_44708730
|
0.086
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr16_+_2039945
|
0.086
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr17_+_64298844
|
0.086
|
NM_002737
|
PRKCA
|
protein kinase C, alpha
|
|
chr9_-_98269480
|
0.085
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr12_+_65672422
|
0.085
|
NM_001031679
NM_001193460
NM_198080
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr10_+_1102741
|
0.085
|
NM_014023
|
WDR37
|
WD repeat domain 37
|
|
chr12_-_22487566
|
0.085
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr20_+_56884749
|
0.085
|
NM_020673
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr8_+_70378858
|
0.084
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr2_-_1748285
|
0.084
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr7_+_117824085
|
0.084
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr5_-_124080773
|
0.084
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr3_+_185000714
|
0.083
|
NM_001242314
NM_001242317
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr2_-_55459698
|
0.082
|
NM_001135598
|
C2orf63
|
chromosome 2 open reading frame 63
|
|
chr5_+_173315313
|
0.082
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr1_+_183605207
|
0.082
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr12_-_29936685
|
0.082
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr8_-_93075190
|
0.081
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_-_95767862
|
0.081
|
NM_001177875
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr13_+_39612440
|
0.081
|
NM_001012754
NM_001017370
|
NHLRC3
|
NHL repeat containing 3
|
|
chr5_+_49961732
|
0.081
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chrX_-_34675366
|
0.081
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr2_-_214016332
|
0.081
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr5_-_39425297
|
0.080
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr12_+_120105656
|
0.080
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr7_+_66386129
|
0.078
|
NM_017994
|
C7orf42
|
chromosome 7 open reading frame 42
|
|
chr22_+_42372763
|
0.078
|
NM_019106
NM_145733
|
SEPT3
|
septin 3
|
|
chr4_-_21699317
|
0.077
|
NM_001035003
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chrX_-_71351750
|
0.077
|
NM_001024455
|
RGAG4
|
retrotransposon gag domain containing 4
|
|
chr6_-_75915566
|
0.077
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr2_+_191745546
|
0.077
|
NM_014905
|
GLS
|
glutaminase
|
|
chr8_+_37654395
|
0.076
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr14_+_74111577
|
0.076
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chrX_-_19988381
|
0.076
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr6_+_52935770
|
0.076
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chrX_+_79591002
|
0.076
|
NM_001170574
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chr2_-_198175494
|
0.075
|
NM_001195144
NM_153697
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr17_-_5095131
|
0.075
|
NM_032530
|
ZNF594
|
zinc finger protein 594
|