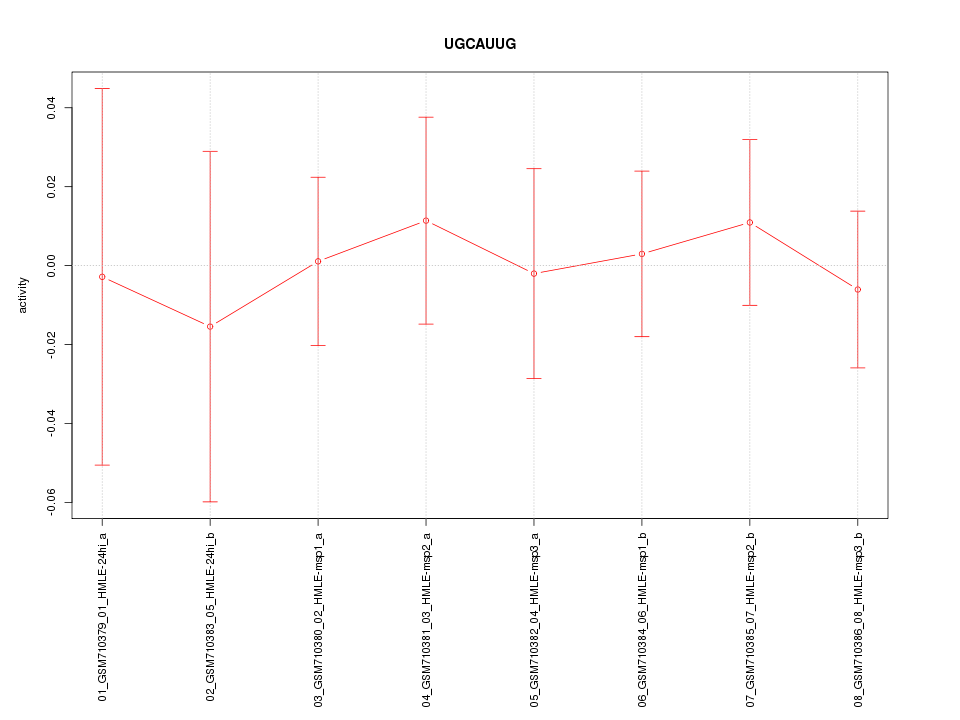
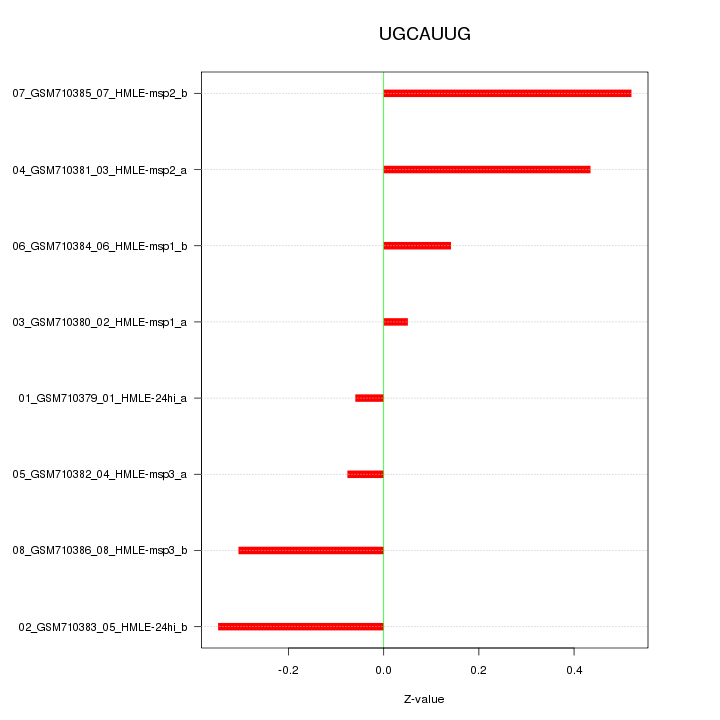
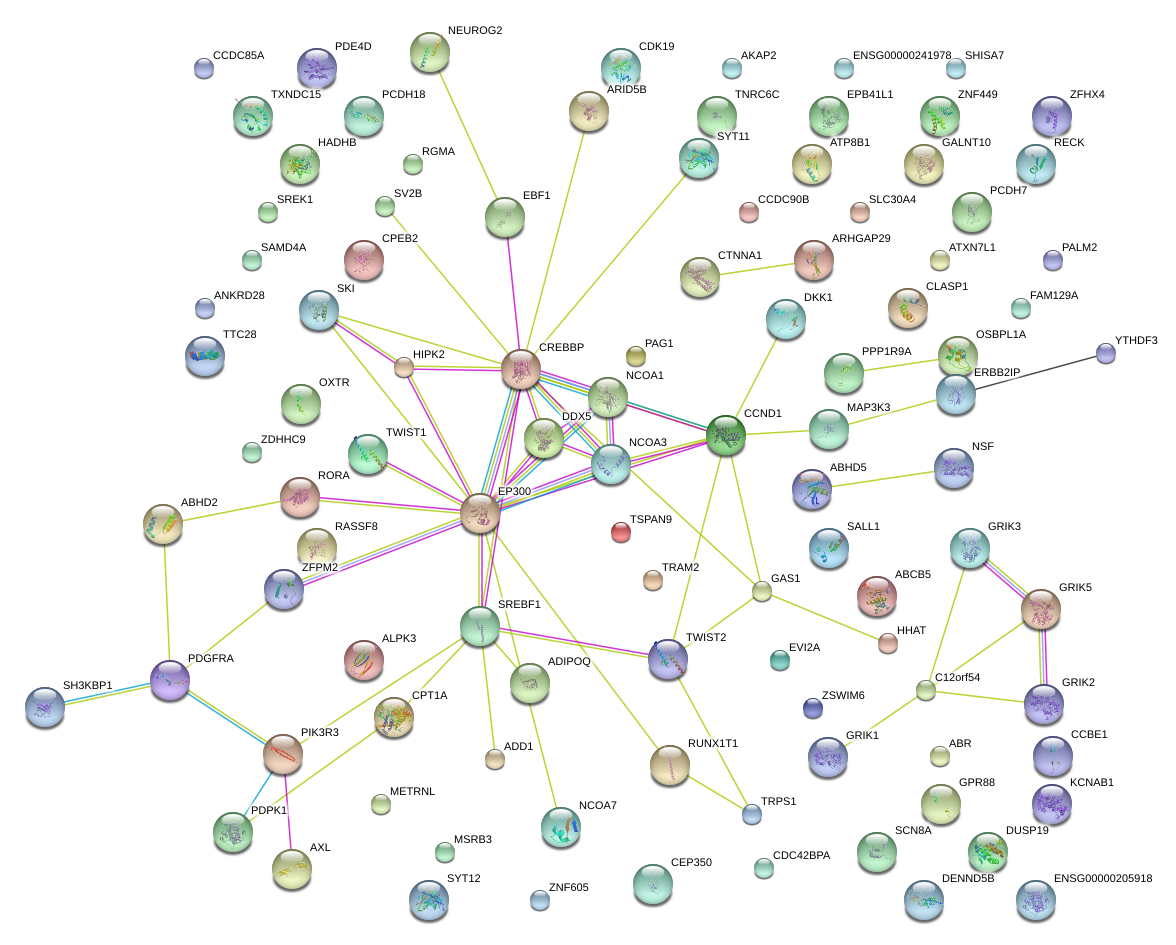
|
chr4_-_138453628
|
0.183
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr1_-_184943673
|
0.132
|
NM_052966
|
FAM129A
|
family with sequence similarity 129, member A
|
|
chr8_+_77593506
|
0.130
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr12_+_65672682
|
0.124
|
NM_001193461
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr12_+_65672422
|
0.123
|
NM_001031679
NM_001193460
NM_198080
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr6_+_101846860
|
0.122
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr9_-_89562103
|
0.115
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr4_+_55095263
|
0.113
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr6_-_52441815
|
0.113
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr3_-_8811263
|
0.101
|
NM_000916
|
OXTR
|
oxytocin receptor
|
|
chr7_-_19157262
|
0.096
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr22_-_29075708
|
0.095
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr5_-_158526698
|
0.091
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr2_+_56411123
|
0.081
|
NM_001080433
|
CCDC85A
|
coiled-coil domain containing 85A
|
|
chr5_+_153570270
|
0.080
|
NM_198321
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10)
|
|
chr8_-_93115453
|
0.078
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr17_+_81037504
|
0.077
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr15_-_93631751
|
0.076
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr4_+_15004297
|
0.076
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr15_-_93632161
|
0.075
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr8_-_93029864
|
0.075
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr15_-_93617091
|
0.075
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr8_-_93111915
|
0.074
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107881
|
0.071
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr15_-_93616891
|
0.071
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr8_-_93107442
|
0.070
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_-_139477437
|
0.067
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr15_-_93616358
|
0.067
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr9_+_36036900
|
0.066
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr16_-_51184507
|
0.066
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr17_+_44668034
|
0.063
|
NM_006178
|
NSF
|
N-ethylmaleimide-sensitive factor
|
|
chr16_-_51185150
|
0.060
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr8_-_93075190
|
0.059
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr17_-_29648631
|
0.058
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr3_+_186560462
|
0.057
|
NM_001177800
NM_004797
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr18_-_55470279
|
0.057
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr3_-_15900928
|
0.057
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr15_+_89631380
|
0.056
|
NM_007011
NM_152924
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr1_-_46598250
|
0.053
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr10_+_54074037
|
0.052
|
NM_012242
|
DKK1
|
dickkopf 1 homolog (Xenopus laevis)
|
|
chr5_+_134209426
|
0.052
|
NM_024715
|
TXNDC15
|
thioredoxin domain containing 15
|
|
chr14_+_55034329
|
0.051
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr12_-_31743822
|
0.050
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr10_+_63661012
|
0.043
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr6_+_126102278
|
0.043
|
NM_001199619
NM_001199620
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr1_+_155829259
|
0.042
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr5_-_58334936
|
0.042
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr18_-_57364643
|
0.041
|
NM_133459
|
CCBE1
|
collagen and calcium binding EGF domains 1
|
|
chr1_+_179923870
|
0.041
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr3_-_15798057
|
0.040
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr8_-_82024277
|
0.039
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr15_+_91643429
|
0.038
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr17_-_1083008
|
0.037
|
NM_021962
|
ABR
|
active BCR-related gene
|
|
chr12_+_26126687
|
0.036
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr15_-_60919642
|
0.036
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_-_60884615
|
0.034
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr16_+_2587951
|
0.033
|
NM_002613
NM_031268
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chrX_-_19817907
|
0.033
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chrX_-_19688991
|
0.033
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr7_-_105319608
|
0.031
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr12_+_26205490
|
0.031
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr17_-_1090562
|
0.031
|
NM_001159746
|
ABR
|
active BCR-related gene
|
|
chr3_-_15839674
|
0.030
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr6_+_126111782
|
0.030
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr2_+_24807345
|
0.029
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr5_-_59783885
|
0.028
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_1012257
|
0.028
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr5_-_58295758
|
0.028
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_138089073
|
0.028
|
NM_001903
|
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa
|
|
chr14_+_55221505
|
0.027
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr8_+_106331146
|
0.027
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr4_-_113437327
|
0.027
|
NM_024019
|
NEUROG2
|
neurogenin 2
|
|
chr15_+_85359910
|
0.027
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr4_+_2845552
|
0.026
|
NM_001119
NM_014189
NM_014190
NM_176801
|
ADD1
|
adducin 1 (alpha)
|
|
chr1_+_2160133
|
0.026
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr2_-_122406994
|
0.026
|
NM_001142273
NM_001142274
NM_001207051
NM_015282
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr17_+_76000248
|
0.025
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr6_+_126240369
|
0.025
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chrX_-_19905665
|
0.025
|
NM_031892
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr16_-_3930120
|
0.024
|
NM_001079846
NM_004380
|
CREBBP
|
CREB binding protein
|
|
chr5_-_59189620
|
0.024
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr20_+_34700257
|
0.023
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr9_+_112887780
|
0.022
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chrX_+_134478671
|
0.021
|
NM_152695
|
ZNF449
|
zinc finger protein 449
|
|
chr11_-_68609358
|
0.021
|
NM_001031847
NM_001876
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver)
|
|
chr7_+_20655244
|
0.020
|
NM_001163941
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr9_+_112810877
|
0.020
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr12_+_3186520
|
0.020
|
NM_001168320
NM_006675
|
TSPAN9
|
tetraspanin 9
|
|
chr20_+_34742656
|
0.020
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr17_-_62502410
|
0.019
|
NM_004396
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5
|
|
chr8_-_116681103
|
0.019
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr3_+_156008775
|
0.019
|
NM_172159
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr1_+_101003692
|
0.019
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chr15_-_45815001
|
0.019
|
NM_013309
|
SLC30A4
|
solute carrier family 30 (zinc transporter), member 4
|
|
chr3_+_43732358
|
0.019
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr11_+_69455837
|
0.018
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr19_+_41725098
|
0.018
|
NM_001699
NM_021913
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr17_+_61699786
|
0.018
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr7_+_94536926
|
0.018
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr20_+_46130600
|
0.018
|
NM_001174087
NM_001174088
NM_006534
NM_181659
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chr12_+_26111951
|
0.017
|
NM_001164748
NM_007211
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr1_-_227505796
|
0.017
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr6_-_111136407
|
0.017
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr17_-_17740309
|
0.017
|
NM_001005291
NM_004176
|
SREBF1
|
sterol regulatory element binding transcription factor 1
|
|
chr5_-_59064437
|
0.016
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_51985016
|
0.016
|
NM_014191
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr5_+_60628085
|
0.015
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr5_-_58652671
|
0.015
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr8_+_64081108
|
0.015
|
NM_152758
|
YTHDF3
|
YTH domain family, member 3
|
|
chr2_+_26467615
|
0.015
|
NM_000183
|
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit
|
|
chr3_+_155860750
|
0.015
|
NM_003471
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr2_+_183943286
|
0.015
|
NM_001142314
NM_080876
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr4_+_30721865
|
0.015
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr5_-_58571916
|
0.014
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chrX_-_128977291
|
0.014
|
NM_001008222
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr5_+_65222381
|
0.014
|
NM_001006600
NM_001253697
NM_001253698
NM_001253699
NM_001253701
NM_018695
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr1_-_94703252
|
0.014
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr5_+_65440662
|
0.013
|
NM_139168
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr5_-_108745569
|
0.013
|
NM_014819
|
PJA2
|
praja ring finger 2
|
|
chr2_+_178257371
|
0.013
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr2_+_149402559
|
0.013
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr15_-_61521478
|
0.012
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_+_79724857
|
0.012
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr20_+_13976014
|
0.012
|
NM_080676
|
MACROD2
|
MACRO domain containing 2
|
|
chr22_-_29196537
|
0.012
|
NM_001079539
NM_005080
|
XBP1
|
X-box binding protein 1
|
|
chr8_-_38386174
|
0.012
|
NM_207412
|
C8orf86
|
chromosome 8 open reading frame 86
|
|
chr6_+_34482646
|
0.012
|
NM_001199583
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr10_+_75757847
|
0.011
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr12_+_111843743
|
0.011
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr15_-_56035176
|
0.011
|
NM_173814
|
PRTG
|
protogenin
|
|
chr3_+_85008031
|
0.011
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr6_-_74230732
|
0.011
|
NM_001402
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr13_-_53024730
|
0.011
|
NM_016075
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae)
|
|
chr12_+_69864065
|
0.011
|
NM_001042555
NM_006654
|
FRS2
|
fibroblast growth factor receptor substrate 2
|
|
chr3_-_64211111
|
0.011
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr6_+_36562089
|
0.010
|
NM_003017
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr10_-_94050799
|
0.010
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr2_-_37193609
|
0.010
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chrX_-_128977719
|
0.010
|
NM_016032
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr12_+_64238511
|
0.010
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr12_+_52056547
|
0.010
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr20_+_31407696
|
0.010
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr6_+_69344939
|
0.009
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr6_+_34433849
|
0.009
|
NM_020804
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr15_+_91643181
|
0.009
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr1_-_119532178
|
0.009
|
NM_152380
|
TBX15
|
T-box 15
|
|
chr18_-_18691765
|
0.009
|
NM_005406
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1
|
|
chr1_+_204485461
|
0.009
|
NM_001204171
NM_001204172
NM_002393
|
MDM4
|
Mdm4 p53 binding protein homolog (mouse)
|
|
chr1_+_26737232
|
0.009
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr20_+_15177503
|
0.009
|
NM_001033087
|
MACROD2
|
MACRO domain containing 2
|
|
chr7_+_155250705
|
0.009
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr16_+_69220940
|
0.008
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr7_-_105516923
|
0.008
|
NM_020725
NM_152749
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr16_-_70472920
|
0.008
|
NM_006927
|
ST3GAL2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chr3_+_155838336
|
0.008
|
NM_172160
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr7_+_105603656
|
0.008
|
NM_152750
|
CDHR3
|
cadherin-related family member 3
|
|
chr17_-_58469577
|
0.008
|
NM_032582
|
USP32
|
ubiquitin specific peptidase 32
|
|
chr12_-_56652118
|
0.008
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr2_-_86564632
|
0.008
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr1_+_22889988
|
0.008
|
NM_001006943
NM_020526
|
EPHA8
|
EPH receptor A8
|
|
chr13_-_110438896
|
0.008
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr4_-_85887543
|
0.008
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr17_+_5031686
|
0.007
|
NM_004505
|
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene)
|
|
chr4_+_40058507
|
0.007
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr2_-_99347588
|
0.007
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chrX_-_24045302
|
0.007
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr12_+_1100352
|
0.007
|
NM_178039
NM_178040
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr21_-_44846927
|
0.007
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr3_+_173116243
|
0.007
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr9_+_130854127
|
0.007
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr6_-_56507585
|
0.006
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr4_+_134070443
|
0.006
|
NM_020815
NM_032961
|
PCDH10
|
protocadherin 10
|
|
chr5_-_40798263
|
0.006
|
NM_006251
NM_206907
|
PRKAA1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit
|
|
chr15_-_23086290
|
0.006
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr9_+_34990266
|
0.006
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr1_+_10270680
|
0.006
|
NM_015074
NM_183416
|
KIF1B
|
kinesin family member 1B
|
|
chr15_-_51914770
|
0.006
|
NM_001174116
NM_001174117
NM_015263
|
DMXL2
|
Dmx-like 2
|
|
chr17_+_53046096
|
0.006
|
NM_178509
|
STXBP4
|
syntaxin binding protein 4
|
|
chr14_+_53196852
|
0.006
|
NM_001130701
NM_145251
|
STYX
|
serine/threonine/tyrosine interacting protein
|
|
chr7_-_126883568
|
0.006
|
NM_000845
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr1_+_118148506
|
0.006
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr10_+_64564480
|
0.005
|
NM_032804
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase
|
|
chr2_+_200775974
|
0.005
|
NM_153689
|
C2orf69
|
chromosome 2 open reading frame 69
|
|
chr7_+_94539209
|
0.005
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr1_+_182992329
|
0.005
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr21_+_38071990
|
0.005
|
NM_005069
NM_009586
|
SIM2
|
single-minded homolog 2 (Drosophila)
|
|
chr5_-_58882274
|
0.005
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_22778008
|
0.005
|
NM_001039481
NM_018638
|
ETNK1
|
ethanolamine kinase 1
|
|
chr14_+_57046490
|
0.005
|
NM_017799
|
C14orf101
|
chromosome 14 open reading frame 101
|
|
chr18_-_21166440
|
0.004
|
NM_000271
|
NPC1
|
Niemann-Pick disease, type C1
|
|
chr1_+_218458628
|
0.004
|
NM_016052
|
RRP15
|
ribosomal RNA processing 15 homolog (S. cerevisiae)
|
|
chr7_+_86974928
|
0.004
|
NM_001143935
NM_001243745
NM_021151
|
CROT
|
carnitine O-octanoyltransferase
|
|
chr8_+_98656329
|
0.004
|
NM_178812
|
MTDH
|
metadherin
|
|
chr3_-_141719188
|
0.004
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr12_-_104531875
|
0.004
|
NM_006166
|
NFYB
|
nuclear transcription factor Y, beta
|
|
chr5_-_160975124
|
0.004
|
NM_000813
NM_021911
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr8_+_117950437
|
0.004
|
NM_001025357
|
C8orf85
|
chromosome 8 open reading frame 85
|
|
chr2_-_86565205
|
0.004
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr9_+_5629110
|
0.004
|
NM_001135920
NM_001206557
NM_020829
|
KIAA1432
|
KIAA1432
|
|
chr5_+_65440004
|
0.003
|
NM_001077199
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|