|
chr4_+_55095263
|
0.650
|
NM_006206
|
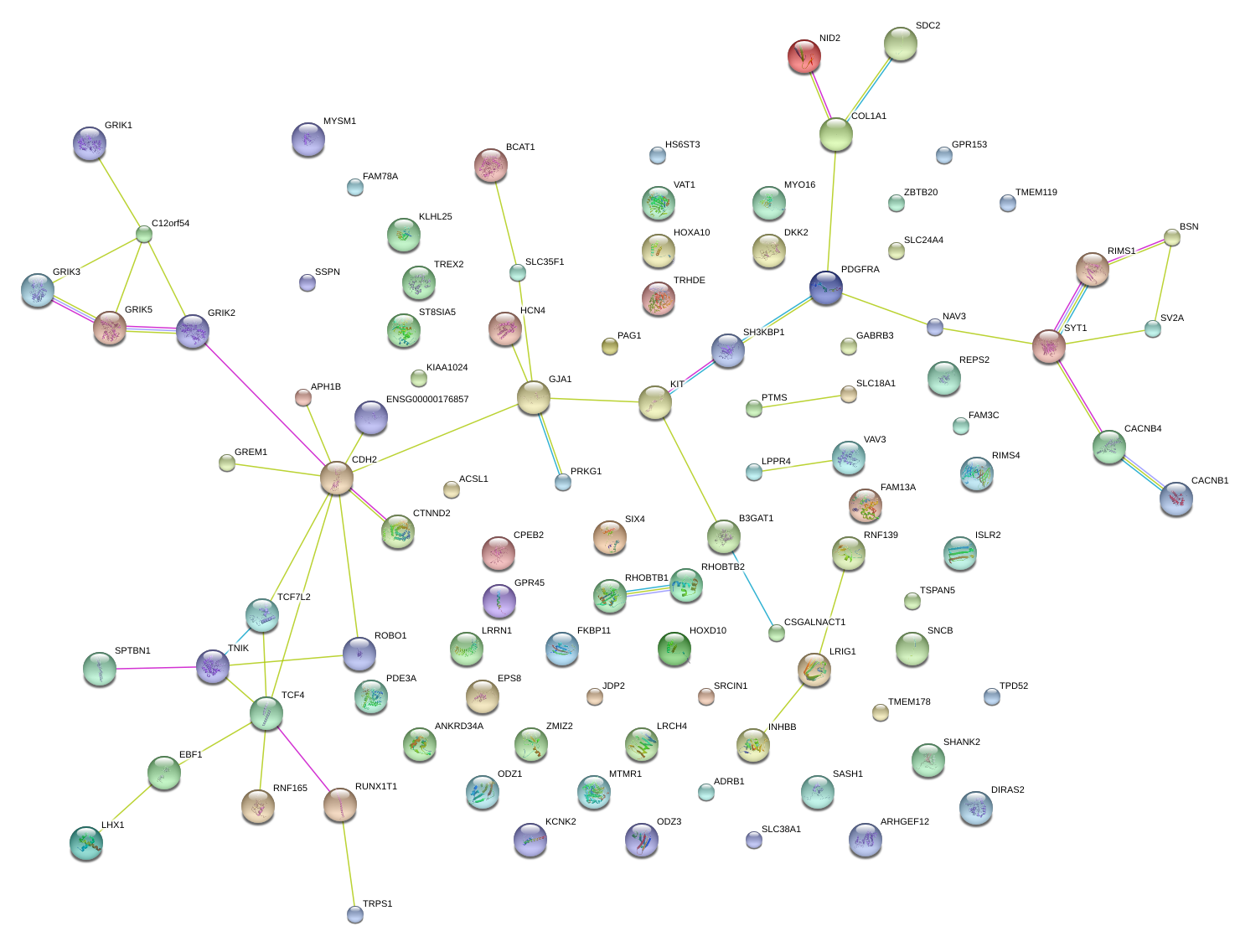
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr8_-_93115453
|
0.512
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr6_+_101846860
|
0.511
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr8_-_93111915
|
0.503
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107881
|
0.492
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93029864
|
0.489
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107442
|
0.474
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93075190
|
0.439
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_+_26348268
|
0.423
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr12_-_25055228
|
0.417
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr12_-_25055966
|
0.416
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr12_-_25102281
|
0.402
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr12_+_26348488
|
0.369
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr6_+_72926462
|
0.359
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_+_72922513
|
0.337
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_+_73076431
|
0.327
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_+_72926144
|
0.326
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_149889362
|
0.319
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr8_+_97505876
|
0.289
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr15_+_33010174
|
0.269
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr1_+_215256559
|
0.253
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr1_+_215178884
|
0.251
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr12_+_72666462
|
0.234
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr11_-_70935807
|
0.230
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr11_-_70507911
|
0.225
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr2_+_39892637
|
0.219
|
NM_001167959
|
TMEM178
|
transmembrane protein 178
|
|
chr2_+_39893021
|
0.214
|
NM_152390
|
TMEM178
|
transmembrane protein 178
|
|
chr8_-_81083660
|
0.199
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr5_-_158526698
|
0.197
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr6_+_148663728
|
0.190
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr8_-_80993009
|
0.188
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr18_-_44336991
|
0.185
|
NM_013305
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chr17_-_48278987
|
0.184
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr4_-_107957452
|
0.169
|
NM_014421
|
DKK2
|
dickkopf 2 homolog (Xenopus laevis)
|
|
chr3_-_66550844
|
0.161
|
NM_015541
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr6_+_72596648
|
0.160
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_145470507
|
0.160
|
NM_001039888
|
ANKRD34A
|
ankyrin repeat domain 34A
|
|
chr12_+_79257772
|
0.158
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr14_+_75898836
|
0.153
|
NM_001135049
|
JDP2
|
Jun dimerization protein 2
|
|
chr12_+_79439432
|
0.152
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr2_+_121103670
|
0.149
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr12_+_79258448
|
0.146
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chrX_+_16964730
|
0.145
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr15_-_86338178
|
0.144
|
NM_022480
|
KLHL25
|
kelch-like 25 (Drosophila)
|
|
chr18_-_25757351
|
0.144
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr3_+_3841079
|
0.142
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr14_+_75894919
|
0.140
|
NM_001135048
NM_130469
|
JDP2
|
Jun dimerization protein 2
|
|
chr8_-_116681103
|
0.136
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr5_-_176057324
|
0.135
|
NM_001001502
NM_003085
|
SNCB
|
synuclein, beta
|
|
chr9_-_93405066
|
0.133
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr1_+_99729847
|
0.133
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr10_-_62703822
|
0.126
|
NM_001242359
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr4_-_89744154
|
0.126
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr6_+_118228688
|
0.124
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr14_+_75894508
|
0.124
|
NM_001135047
|
JDP2
|
Jun dimerization protein 2
|
|
chr14_+_92790151
|
0.122
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr5_-_11903964
|
0.120
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr6_+_121756744
|
0.120
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr11_-_134281725
|
0.116
|
NM_018644
NM_054025
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P)
|
|
chr1_-_108507474
|
0.116
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr14_+_92788924
|
0.116
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr3_-_114102488
|
0.115
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_-_114790221
|
0.115
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_-_46663207
|
0.114
|
NM_001077484
NM_030674
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr3_-_114343791
|
0.112
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_-_114478117
|
0.110
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_+_20522178
|
0.108
|
NM_000921
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr20_-_43438881
|
0.106
|
NM_001205317
NM_182970
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|
|
chr1_-_108231122
|
0.106
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr3_-_79068542
|
0.101
|
NM_001145845
NM_133631
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr17_-_36762105
|
0.101
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr14_+_92789536
|
0.097
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr12_-_108991893
|
0.096
|
NM_181724
|
TMEM119
|
transmembrane protein 119
|
|
chr2_+_105858199
|
0.096
|
NM_007227
|
GPR45
|
G protein-coupled receptor 45
|
|
chr3_-_114343052
|
0.095
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr4_+_15004297
|
0.095
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr15_-_27018917
|
0.095
|
NM_021912
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr11_+_120207617
|
0.094
|
NM_001198665
NM_015313
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12
|
|
chr15_-_26961911
|
0.093
|
NM_001191320
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr7_+_44788453
|
0.092
|
NM_031449
|
ZMIZ2
|
zinc finger, MIZ-type containing 2
|
|
chrX_-_19817907
|
0.091
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr8_-_19540093
|
0.090
|
NM_018371
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1
|
|
chr18_+_43914186
|
0.090
|
NM_152470
|
RNF165-IT1
RNF165
|
RNF165 intronic transcript 1 (non-protein coding)
ring finger protein 165
|
|
chr13_+_109281569
|
0.090
|
NM_001198950
|
MYO16
|
myosin XVI
|
|
chrX_-_19905665
|
0.086
|
NM_031892
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr2_-_152955208
|
0.086
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr17_+_35294771
|
0.085
|
NM_005568
|
LHX1
|
LIM homeobox 1
|
|
chr3_-_171177851
|
0.085
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr4_-_99579540
|
0.084
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr9_-_134151905
|
0.084
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr10_+_52834233
|
0.084
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr15_-_27018216
|
0.083
|
NM_000814
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr13_+_109248499
|
0.081
|
NM_015011
|
MYO16
|
myosin XVI
|
|
chr12_-_15942350
|
0.080
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr10_+_115803805
|
0.080
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr13_+_96743092
|
0.079
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr4_-_89978117
|
0.078
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr18_-_53255734
|
0.078
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr12_+_104697460
|
0.078
|
NM_001008394
|
EID3
|
EP300 interacting inhibitor of differentiation 3
|
|
chr12_+_78225068
|
0.078
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr10_+_52750904
|
0.077
|
NM_001098512
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr1_-_59165735
|
0.077
|
NM_001085487
|
MYSM1
|
Myb-like, SWIRM and MPN domains 1
|
|
chr4_+_55523906
|
0.076
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr2_-_152830448
|
0.075
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr2_-_152955502
|
0.075
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr7_+_44795786
|
0.074
|
NM_174929
|
ZMIZ2
|
zinc finger, MIZ-type containing 2
|
|
chr15_+_63569748
|
0.073
|
NM_001145646
NM_031301
|
APH1B
|
anterior pharynx defective 1 homolog B (C. elegans)
|
|
chr8_-_82024277
|
0.071
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr7_-_27213873
|
0.071
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr2_+_54785453
|
0.070
|
NM_178313
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr3_-_79817058
|
0.070
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr15_+_74421986
|
0.070
|
NM_001130137
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr15_+_74422589
|
0.070
|
NM_001130138
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr3_+_49591897
|
0.070
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr1_-_6321034
|
0.069
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr17_-_41174431
|
0.069
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr15_+_79724857
|
0.068
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr15_+_74421698
|
0.067
|
NM_001130136
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chrX_+_149861830
|
0.066
|
NM_003828
|
MTMR1
|
myotubularin related protein 1
|
|
chr4_-_185747183
|
0.066
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr7_-_121036355
|
0.066
|
NM_001040020
NM_014888
|
FAM3C
|
family with sequence similarity 3, member C
|
|
chrX_-_19688991
|
0.066
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr8_-_19460005
|
0.065
|
NM_001130518
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1
|
|
chr8_+_125486949
|
0.065
|
NM_007218
|
RNF139
|
ring finger protein 139
|
|
chr14_-_61190458
|
0.064
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr15_+_74422742
|
0.064
|
NM_020851
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr15_-_73661604
|
0.064
|
NM_005477
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chr4_+_183245136
|
0.064
|
NM_001080477
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr14_-_52535945
|
0.063
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr6_-_105584542
|
0.063
|
NM_007073
|
BVES
|
blood vessel epicardial substance
|
|
chr1_-_119532178
|
0.063
|
NM_152380
|
TBX15
|
T-box 15
|
|
chr1_-_21978287
|
0.063
|
NM_001145658
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr12_-_56615679
|
0.062
|
NM_005785
NM_194358
NM_194359
|
RNF41
|
ring finger protein 41
|
|
chr3_+_154798078
|
0.062
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_+_154797435
|
0.062
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr19_-_41196419
|
0.062
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr3_+_154797910
|
0.062
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_-_21995793
|
0.062
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr3_+_154797644
|
0.062
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr5_+_49961732
|
0.062
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr6_-_105584220
|
0.061
|
NM_147147
|
BVES
|
blood vessel epicardial substance
|
|
chr16_-_52581713
|
0.060
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr15_-_50411405
|
0.060
|
NM_024837
|
ATP8B4
|
ATPase, class I, type 8B, member 4
|
|
chr6_-_105584982
|
0.060
|
NM_001199563
|
BVES
|
blood vessel epicardial substance
|
|
chr16_-_52580805
|
0.059
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr9_-_123476578
|
0.059
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr3_-_14583587
|
0.059
|
NM_001080423
|
GRIP2
|
glutamate receptor interacting protein 2
|
|
chr1_+_113051369
|
0.059
|
NM_024494
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr11_-_78052925
|
0.058
|
NM_012296
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr3_+_11034419
|
0.058
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr17_+_1182956
|
0.057
|
NM_172367
|
TUSC5
|
tumor suppressor candidate 5
|
|
chr10_+_72575623
|
0.057
|
NM_003901
|
SGPL1
|
sphingosine-1-phosphate lyase 1
|
|
chr17_-_76356153
|
0.057
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr12_-_54673901
|
0.056
|
NM_012117
|
CBX5
|
chromobox homolog 5
|
|
chr4_+_39046198
|
0.056
|
NM_001007075
NM_001171654
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr6_-_36515245
|
0.056
|
NM_007271
|
STK38
|
serine/threonine kinase 38
|
|
chr14_+_78870092
|
0.056
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr6_-_128841432
|
0.056
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr8_-_145559831
|
0.056
|
NM_031309
|
SCRT1
|
scratch homolog 1, zinc finger protein (Drosophila)
|
|
chr11_-_78128852
|
0.055
|
NM_080491
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr12_-_56615484
|
0.055
|
NM_001242826
|
RNF41
|
ring finger protein 41
|
|
chr9_+_34958171
|
0.054
|
NM_015297
|
KIAA1045
|
KIAA1045
|
|
chr5_+_49962771
|
0.054
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr22_+_24666749
|
0.054
|
NM_001145468
NM_015330
|
SPECC1L
|
sperm antigen with calponin homology and coiled-coil domains 1-like
|
|
chr4_-_6565326
|
0.054
|
NM_001206994
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr4_-_6557404
|
0.054
|
NM_001206995
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr22_+_25202135
|
0.054
|
NM_001039948
NM_001098497
NM_001098498
NM_133454
|
SGSM1
|
small G protein signaling modulator 1
|
|
chr12_-_22487566
|
0.053
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr11_+_118401755
|
0.053
|
NM_001144034
NM_001144035
NM_001144036
NM_001144037
NM_001144038
NM_032780
|
TMEM25
|
transmembrane protein 25
|
|
chr10_+_112257624
|
0.052
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr4_+_52709165
|
0.052
|
NM_001040402
NM_015115
|
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr21_+_35445808
|
0.051
|
NM_032476
NM_006933
|
MRPS6
SLC5A3
|
mitochondrial ribosomal protein S6
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3
|
|
chr6_+_136172802
|
0.051
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chrX_+_51075082
|
0.051
|
NM_153183
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10
|
|
chr4_+_39063851
|
0.051
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr1_+_9352931
|
0.050
|
NM_025106
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr9_-_127905715
|
0.050
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr21_-_34100228
|
0.049
|
NM_001160302
NM_001160306
|
SYNJ1
|
synaptojanin 1
|
|
chr12_-_71182699
|
0.048
|
NM_001207015
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr17_+_42264300
|
0.048
|
NM_001076674
NM_024107
|
TMUB2
|
transmembrane and ubiquitin-like domain containing 2
|
|
chr12_+_94542239
|
0.048
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr1_+_113010039
|
0.047
|
NM_004185
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr2_+_45878912
|
0.046
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr3_-_69435429
|
0.046
|
NM_015123
|
FRMD4B
|
FERM domain containing 4B
|
|
chr4_+_95679075
|
0.046
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr1_+_28832454
|
0.045
|
NM_001048199
|
RCC1
|
regulator of chromosome condensation 1
|
|
chr4_-_6422844
|
0.045
|
NM_001206996
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr8_+_22224997
|
0.044
|
NM_001135153
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr3_+_151985828
|
0.044
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr7_-_71877359
|
0.044
|
NM_031468
|
CALN1
|
calneuron 1
|
|
chr19_-_55954229
|
0.044
|
NM_001145176
|
SHISA7
|
shisa homolog 7 (Xenopus laevis)
|
|
chr7_+_106685007
|
0.044
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr14_+_85996454
|
0.044
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr17_+_60556247
|
0.043
|
NM_001112707
NM_006852
|
TLK2
|
tousled-like kinase 2
|
|
chr4_+_38665587
|
0.043
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr3_-_53079285
|
0.043
|
NM_001005159
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr1_-_57045235
|
0.043
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr7_+_155089483
|
0.043
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr21_-_34100316
|
0.043
|
NM_003895
NM_203446
|
SYNJ1
|
synaptojanin 1
|
|
chr6_-_37665631
|
0.042
|
NM_153487
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|