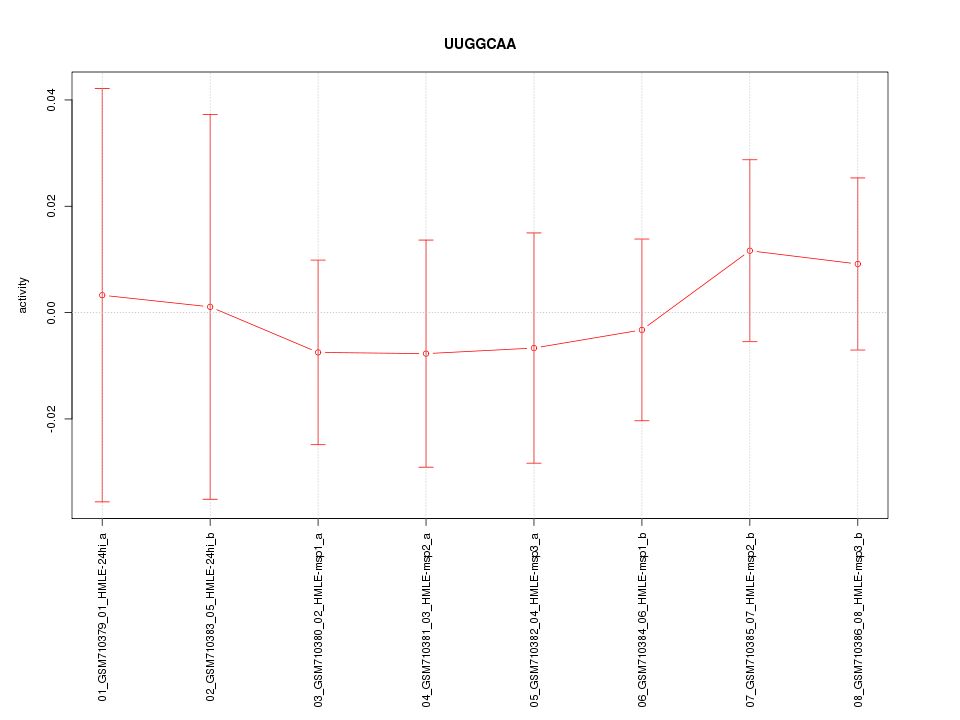
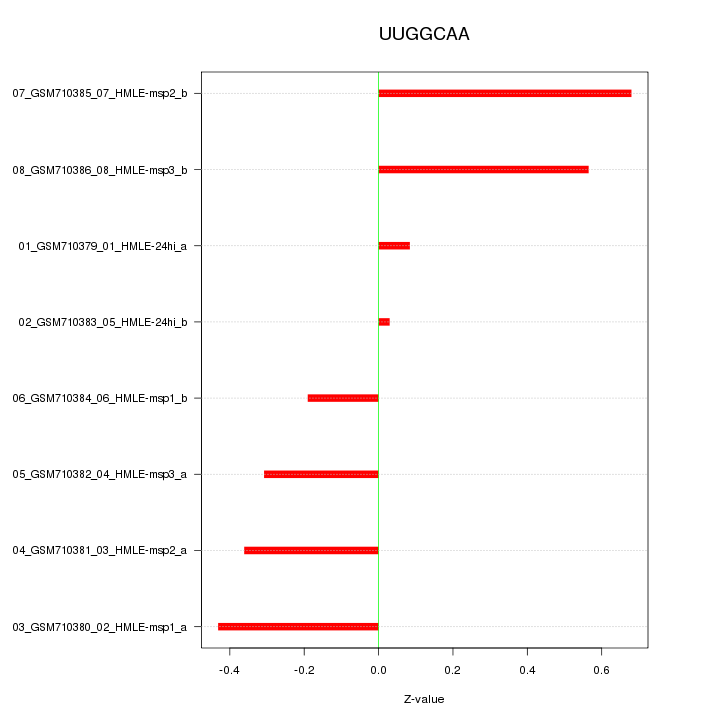
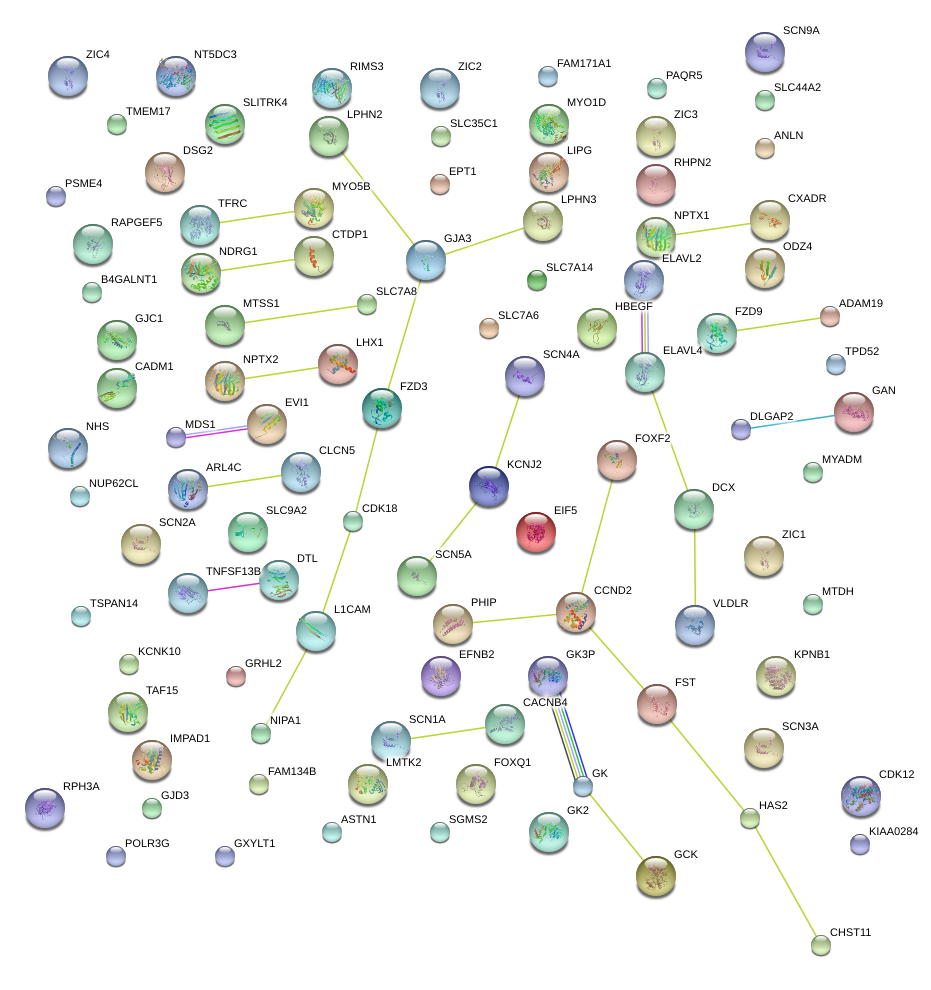
|
chr18_+_47088400
|
0.197
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr9_+_2621740
|
0.170
|
NM_001018056
NM_003383
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr17_+_68165660
|
0.159
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr19_+_54372659
|
0.139
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_+_54369610
|
0.133
|
NM_001020820
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_+_54371125
|
0.126
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr5_-_139726173
|
0.122
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr7_+_97736118
|
0.119
|
NM_014916
|
LMTK2
|
lemur tyrosine kinase 2
|
|
chr2_+_26568900
|
0.111
|
NM_033505
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr1_+_82266081
|
0.109
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chrX_+_49832214
|
0.099
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr8_-_122653493
|
0.097
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr12_+_4382882
|
0.093
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr11_-_79151694
|
0.090
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr5_-_16617085
|
0.087
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr5_-_16509106
|
0.086
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr2_-_235405692
|
0.086
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr6_-_79787939
|
0.086
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr2_-_166060552
|
0.084
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chrX_+_49687212
|
0.083
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr12_-_58026846
|
0.082
|
NM_001478
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1
|
|
chr9_-_23826062
|
0.081
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chrX_-_110654373
|
0.080
|
NM_000555
|
DCX
|
doublecortin
|
|
chr3_-_195808958
|
0.077
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr8_-_80993009
|
0.076
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr5_+_52776263
|
0.075
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr17_-_42907606
|
0.075
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr5_+_89770680
|
0.075
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr11_+_45826640
|
0.075
|
NM_018389
|
SLC35C1
|
solute carrier family 35, member C1
|
|
chr4_+_108745718
|
0.075
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr9_-_23821477
|
0.074
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr8_+_28351694
|
0.074
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr16_+_81348564
|
0.073
|
NM_022041
|
GAN
|
gigaxonin
|
|
chr3_-_195808997
|
0.073
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr11_+_45825622
|
0.072
|
NM_001145265
NM_001145266
|
SLC35C1
|
solute carrier family 35, member C1
|
|
chrX_+_17393537
|
0.071
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chrX_+_17653412
|
0.071
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr17_-_42908178
|
0.070
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr13_-_107187312
|
0.069
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr8_-_81083660
|
0.069
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr6_+_1390068
|
0.069
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr9_-_23821842
|
0.069
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr2_-_62733475
|
0.068
|
NM_198276
|
TMEM17
|
transmembrane protein 17
|
|
chr1_-_177133950
|
0.067
|
NM_004319
NM_207108
|
ASTN1
|
astrotactin 1
|
|
chr3_-_168865521
|
0.067
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr15_+_69591272
|
0.067
|
NM_017705
|
PAQR5
|
progestin and adipoQ receptor family member V
|
|
chr8_+_1449531
|
0.067
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr19_-_33555757
|
0.067
|
NM_033103
|
RHPN2
|
rhophilin, Rho GTPase binding protein 2
|
|
chr11_-_115375065
|
0.067
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr10_+_82213892
|
0.066
|
NM_001128309
NM_030927
|
TSPAN14
|
tetraspanin 14
|
|
chr2_-_167232464
|
0.066
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr13_+_100633977
|
0.065
|
NM_007129
|
ZIC2
|
Zic family member 2
|
|
chr15_+_69606741
|
0.065
|
NM_001104554
|
PAQR5
|
progestin and adipoQ receptor family member V
|
|
chr7_+_98246595
|
0.065
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chrX_-_110655384
|
0.065
|
NM_178151
NM_001195553
NM_178152
NM_178153
|
DCX
|
doublecortin
|
|
chr7_-_22396511
|
0.065
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr17_-_31203889
|
0.064
|
NM_015194
|
MYO1D
|
myosin ID
|
|
chr2_-_54197786
|
0.064
|
NM_014614
|
PSME4
|
proteasome (prosome, macropain) activator subunit 4
|
|
chr21_+_18885104
|
0.064
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr3_-_168864066
|
0.064
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chrX_-_142722869
|
0.064
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr8_-_134309448
|
0.064
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chrX_-_106449543
|
0.064
|
NM_017681
|
NUP62CL
|
nucleoporin 62kDa C-terminal like
|
|
chr2_+_103236147
|
0.064
|
NM_003048
|
SLC9A2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2
|
|
chr12_+_113229548
|
0.063
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr8_-_57906423
|
0.063
|
NM_017813
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr4_+_108814419
|
0.062
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr12_-_42538432
|
0.062
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr5_-_157002739
|
0.062
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr14_-_23652840
|
0.061
|
NM_012244
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr1_-_41131323
|
0.060
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr16_+_68298326
|
0.060
|
NM_001076785
NM_003983
|
SLC7A6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6
|
|
chr8_+_102504661
|
0.060
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr14_+_105331581
|
0.059
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr3_-_168864325
|
0.059
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr17_+_34136437
|
0.059
|
NM_003487
NM_139215
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa
|
|
chr10_-_15413054
|
0.059
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr17_+_35294771
|
0.059
|
NM_005568
|
LHX1
|
LIM homeobox 1
|
|
chr18_+_29078026
|
0.058
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr18_-_47721165
|
0.058
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr18_+_77441429
|
0.058
|
NM_001202504
|
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1
|
|
chr8_-_125740650
|
0.058
|
NM_014751
|
MTSS1
|
metastasis suppressor 1
|
|
chr14_+_103800492
|
0.058
|
NM_001969
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr14_-_88737160
|
0.057
|
NM_138318
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chrX_+_30671456
|
0.057
|
NM_000167
NM_001128127
NM_001205019
NM_203391
|
GK
|
glycerol kinase
|
|
chr14_-_23623493
|
0.057
|
NM_182728
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chrX_-_153141398
|
0.057
|
NM_000425
NM_001143963
NM_024003
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr2_-_152955502
|
0.056
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr14_-_81405883
|
0.055
|
NM_152446
|
CEP128
|
centrosomal protein 128kDa
|
|
chr14_-_88793250
|
0.055
|
NM_021161
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr1_+_212208918
|
0.055
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr12_-_104234968
|
0.054
|
NM_001031701
|
NT5DC3
|
5'-nucleotidase domain containing 3
|
|
chr7_+_36429388
|
0.054
|
NM_018685
|
ANLN
|
anillin, actin binding protein
|
|
chr15_-_23086842
|
0.054
|
NM_001142275
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr17_-_78450247
|
0.053
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr3_-_170303774
|
0.052
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (orphan transporter), member 14
|
|
chr1_+_205473683
|
0.052
|
NM_002596
NM_212502
NM_212503
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr17_+_37617688
|
0.051
|
NM_015083
NM_016507
|
CDK12
|
cyclin-dependent kinase 12
|
|
chr6_+_1312674
|
0.051
|
NM_033260
|
FOXQ1
|
forkhead box Q1
|
|
chr12_+_104850641
|
0.050
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr8_+_98656329
|
0.050
|
NM_178812
|
MTDH
|
metadherin
|
|
chr17_+_45727274
|
0.050
|
NM_002265
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr13_-_20735182
|
0.050
|
NM_021954
|
GJA3
|
gap junction protein, alpha 3, 46kDa
|
|
chr19_+_10713055
|
0.049
|
NM_001145056
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr15_-_91537724
|
0.049
|
NM_003981
NM_199413
NM_199414
|
PRC1
|
protein regulator of cytokinesis 1
|
|
chr19_+_10736167
|
0.049
|
NM_020428
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr18_+_29027731
|
0.049
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr14_+_71374111
|
0.049
|
NM_014982
|
PCNX
|
pecanex homolog (Drosophila)
|
|
chrX_-_142722318
|
0.049
|
NM_001184750
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr4_-_80994309
|
0.049
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr1_+_160370363
|
0.049
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr1_-_68962726
|
0.048
|
NM_001114120
NM_017779
|
DEPDC1
|
DEP domain containing 1
|
|
chr8_-_11324184
|
0.047
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chrX_-_142723921
|
0.047
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr15_+_94841429
|
0.047
|
NM_001159643
NM_018349
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr2_-_152955208
|
0.047
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr15_-_23086290
|
0.047
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr20_+_361272
|
0.046
|
NM_021158
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr12_-_94853696
|
0.046
|
NM_001042399
NM_016122
|
CCDC41
|
coiled-coil domain containing 41
|
|
chr18_+_23806385
|
0.046
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr4_+_95679075
|
0.046
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr5_+_154092461
|
0.045
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr14_+_56046796
|
0.045
|
NM_001079521
NM_001079522
NM_004986
NM_182926
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr11_+_57435458
|
0.045
|
NM_015457
|
ZDHHC5
|
zinc finger, DHHC-type containing 5
|
|
chr2_-_152830448
|
0.045
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr12_-_80307690
|
0.045
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr22_+_41488607
|
0.045
|
NM_001429
|
EP300
|
E1A binding protein p300
|
|
chr12_+_32654976
|
0.045
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr18_+_77439798
|
0.044
|
NM_004715
NM_048368
|
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1
|
|
chr2_-_101034049
|
0.044
|
NM_004854
|
CHST10
|
carbohydrate sulfotransferase 10
|
|
chr12_-_80329234
|
0.044
|
NM_001143885
NM_001244990
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr3_+_150804675
|
0.044
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr14_-_88789531
|
0.044
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr20_-_50384899
|
0.044
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr5_+_50678957
|
0.043
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr7_+_18548899
|
0.043
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr6_+_43211221
|
0.043
|
NM_032538
|
TTBK1
|
tau tubulin kinase 1
|
|
chr10_+_71561643
|
0.043
|
NM_001130103
NM_080798
NM_080800
NM_080801
NM_080802
NM_080805
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr1_+_70225857
|
0.042
|
NM_020794
|
LRRC7
|
leucine rich repeat containing 7
|
|
chr6_+_135502445
|
0.042
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr1_-_55352920
|
0.042
|
NM_014762
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr12_+_5019048
|
0.042
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr7_-_112726143
|
0.042
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr1_+_114447906
|
0.041
|
NM_022836
|
DCLRE1B
|
DNA cross-link repair 1B
|
|
chr11_+_844062
|
0.040
|
NM_001025234
|
TSPAN4
|
tetraspanin 4
|
|
chr9_-_36276930
|
0.040
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr17_+_57232859
|
0.040
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr7_+_18126543
|
0.040
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr11_-_16424391
|
0.040
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr11_+_844445
|
0.039
|
NM_001025237
|
TSPAN4
|
tetraspanin 4
|
|
chr12_-_93323088
|
0.039
|
NM_003566
|
EEA1
|
early endosome antigen 1
|
|
chr1_+_27153130
|
0.039
|
NM_032283
|
ZDHHC18
|
zinc finger, DHHC-type containing 18
|
|
chr18_+_48405426
|
0.039
|
NM_001168335
NM_002396
|
ME2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial
|
|
chr5_+_86564727
|
0.039
|
NM_022650
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr11_+_111895537
|
0.039
|
NM_001931
|
DLAT
|
dihydrolipoamide S-acetyltransferase
|
|
chr2_-_68479481
|
0.039
|
NM_000945
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha
|
|
chr5_-_39074494
|
0.039
|
NM_152756
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr11_+_842821
|
0.039
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chr1_-_150947456
|
0.039
|
NM_022075
NM_181746
|
CERS2
|
ceramide synthase 2
|
|
chr1_+_22778343
|
0.039
|
NM_001083621
NM_014870
|
ZBTB40
|
zinc finger and BTB domain containing 40
|
|
chr11_-_16497917
|
0.038
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr3_-_169381417
|
0.038
|
NM_001205194
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr8_-_130951996
|
0.038
|
NM_016623
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr1_+_89149836
|
0.037
|
NM_006256
|
PKN2
|
protein kinase N2
|
|
chr21_-_15755439
|
0.037
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr7_-_82073020
|
0.037
|
NM_000722
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1
|
|
chr12_+_107712151
|
0.037
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr20_+_9198036
|
0.037
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr3_-_27498244
|
0.037
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr1_-_54304107
|
0.037
|
NM_001168551
NM_018087
|
TMEM48
|
transmembrane protein 48
|
|
chr2_-_55646946
|
0.037
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr22_+_45560529
|
0.037
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr2_+_235860616
|
0.037
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr15_-_43398168
|
0.036
|
NM_174916
|
UBR1
|
ubiquitin protein ligase E3 component n-recognin 1
|
|
chr12_-_80328926
|
0.036
|
NM_002480
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr20_+_9288446
|
0.036
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr22_+_26565424
|
0.036
|
NM_001184773
NM_001184774
NM_001184775
NM_001184776
NM_001184777
NM_021115
|
SEZ6L
|
seizure related 6 homolog (mouse)-like
|
|
chr1_-_234614848
|
0.035
|
NM_005646
|
TARBP1
|
TAR (HIV-1) RNA binding protein 1
|
|
chr2_+_220408742
|
0.035
|
NM_001005209
|
TMEM198
|
transmembrane protein 198
|
|
chr12_+_107974633
|
0.035
|
NM_001017523
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr7_+_7222142
|
0.034
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr7_-_127670995
|
0.034
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr10_-_62149487
|
0.034
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr14_+_64682671
|
0.034
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr2_-_102003923
|
0.034
|
NM_153836
|
CREG2
|
cellular repressor of E1A-stimulated genes 2
|
|
chr7_-_111846384
|
0.034
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr8_+_125486949
|
0.033
|
NM_007218
|
RNF139
|
ring finger protein 139
|
|
chr8_+_101170231
|
0.033
|
NM_172218
|
SPAG1
|
sperm associated antigen 1
|
|
chr12_+_52345437
|
0.033
|
NM_004302
NM_020328
|
ACVR1B
|
activin A receptor, type IB
|
|
chr3_+_14444083
|
0.033
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr6_+_50786438
|
0.033
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr5_+_86564044
|
0.033
|
NM_002890
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr10_-_131762017
|
0.033
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr8_+_124780742
|
0.032
|
NM_144963
|
FAM91A1
|
family with sequence similarity 91, member A1
|
|
chr9_+_131218678
|
0.032
|
NM_153433
NM_153440
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr22_-_46933066
|
0.032
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr11_-_47516072
|
0.032
|
NM_198700
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr10_-_61900659
|
0.032
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_-_184724021
|
0.032
|
NM_025191
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3
|
|
chr8_+_101170510
|
0.032
|
NM_003114
|
SPAG1
|
sperm associated antigen 1
|