|
chr14_-_60337350
|
0.474
|
NM_021136
|
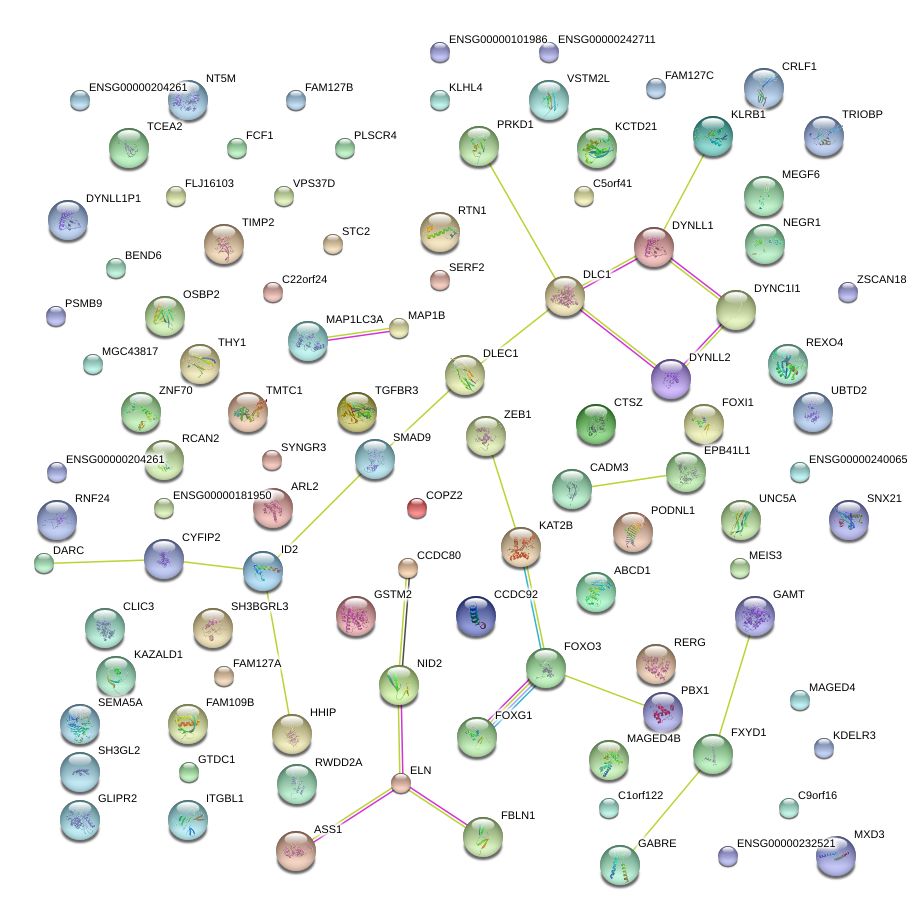
RTN1
|
reticulon 1
|
|
chr11_-_1769086
|
0.467
|
|
IFITM10
|
interferon induced transmembrane protein 10
|
|
chr5_+_156693051
|
0.368
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr1_-_3528058
|
0.357
|
NM_001409
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr5_-_9546157
|
0.347
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr12_-_124457099
|
0.303
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr5_-_172756505
|
0.297
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr16_+_2039945
|
0.290
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr13_+_102104941
|
0.278
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr19_-_18717631
|
0.264
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr2_+_8822112
|
0.262
|
NM_002166
|
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein
|
|
chr4_+_145567147
|
0.259
|
NM_022475
|
HHIP
|
hedgehog interacting protein
|
|
chr1_+_159141376
|
0.245
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chrX_+_51927918
|
0.244
|
NM_001098800
NM_001242362
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr7_+_73082320
|
0.243
|
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chrX_-_51812249
|
0.237
|
NM_001098800
NM_001242362
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr5_+_71403040
|
0.233
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr22_+_45898690
|
0.226
|
NM_001996
NM_006485
NM_006486
NM_006487
|
FBLN1
|
fibulin 1
|
|
chr9_-_139890984
|
0.222
|
NM_004669
|
CLIC3
|
chloride intracellular channel 3
|
|
chr19_-_47922468
|
0.208
|
|
MEIS3
|
Meis homeobox 3
|
|
chr20_+_33146500
|
0.205
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr20_+_34742656
|
0.200
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr1_-_92351476
|
0.199
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr14_+_96342728
|
0.194
|
|
LOC100507043
|
uncharacterized LOC100507043
|
|
chr10_+_31608097
|
0.193
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr5_+_180688174
|
0.190
|
|
|
|
|
chr19_-_18717577
|
0.189
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr19_+_35629727
|
0.188
|
NM_021902
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr6_+_32821924
|
0.185
|
NM_002800
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2)
|
|
chr17_-_76921453
|
0.182
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr6_+_83903031
|
0.179
|
NM_033411
|
RWDD2A
|
RWD domain containing 2A
|
|
chr17_-_76921205
|
0.176
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr17_-_79008372
|
0.171
|
|
FLJ90757
|
uncharacterized LOC440465
|
|
chr1_-_72748147
|
0.168
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chrX_-_151143096
|
0.167
|
NM_004961
|
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon
|
|
chr20_-_3996035
|
0.166
|
NM_007219
NM_001134337
NM_001134338
|
RNF24
|
ring finger protein 24
|
|
chr7_+_73442486
|
0.163
|
|
ELN
|
elastin
|
|
chr1_-_38218573
|
0.163
|
|
EPHA10
|
EPH receptor A10
|
|
chr17_-_46115135
|
0.161
|
NM_016429
|
COPZ2
|
coatomer protein complex, subunit zeta 2
|
|
chr6_-_46459710
|
0.160
|
NM_001251973
|
RCAN2
|
regulator of calcineurin 2
|
|
chr5_+_169532900
|
0.154
|
NM_012188
NM_144769
|
FOXI1
|
forkhead box I1
|
|
chr17_-_5404304
|
0.153
|
NM_001162371
|
LOC728392
|
uncharacterized LOC728392
|
|
chr3_+_20081853
|
0.153
|
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chrX_+_152990322
|
0.152
|
NM_000033
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr1_+_159175048
|
0.150
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr3_-_145968958
|
0.149
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr15_+_44069272
|
0.149
|
NM_001199877
|
SERF2
|
small EDRK-rich factor 2
|
|
chr6_-_46459010
|
0.145
|
NM_001251974
|
RCAN2
|
regulator of calcineurin 2
|
|
chr20_+_54933982
|
0.144
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr3_-_145968819
|
0.144
|
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr20_+_44462395
|
0.143
|
NM_001042632
NM_001042633
NM_033421
NM_152897
|
SNX21
|
sorting nexin family member 21
|
|
chr17_-_76921076
|
0.143
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr19_-_1401474
|
0.142
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr3_+_20081523
|
0.142
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr11_-_77899640
|
0.142
|
NM_001029859
|
KCTD21
|
potassium channel tetramerisation domain containing 21
|
|
chr3_-_187871816
|
0.141
|
|
LPP-AS2
|
LPP antisense RNA 2 (non-protein coding)
|
|
chr20_+_36531548
|
0.140
|
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr9_+_36136710
|
0.139
|
NM_022343
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr2_-_25016214
|
0.138
|
NM_001013663
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1
|
|
chr9_+_130922471
|
0.137
|
NM_024112
|
C9orf16
|
chromosome 9 open reading frame 16
|
|
chr14_-_52535945
|
0.136
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr5_+_176237435
|
0.135
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr20_+_62688438
|
0.135
|
NM_198723
|
TCEA2
|
transcription elongation factor A (SII), 2
|
|
chrX_+_86772714
|
0.133
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr10_+_102820914
|
0.132
|
NM_030929
|
KAZALD1
|
Kazal-type serine peptidase inhibitor domain 1
|
|
chr22_-_24092951
|
0.131
|
NM_021916
|
ZNF70
|
zinc finger protein 70
|
|
chr19_-_47922391
|
0.130
|
|
MEIS3
|
Meis homeobox 3
|
|
chr12_-_15374290
|
0.130
|
NM_001190726
NM_032918
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor
|
|
chr6_+_108881010
|
0.129
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr1_+_110210643
|
0.128
|
NM_000848
NM_001142368
|
GSTM2
|
glutathione S-transferase mu 2 (muscle)
|
|
chr5_+_172484353
|
0.128
|
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr12_-_29936685
|
0.127
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr20_-_57582152
|
0.127
|
|
CTSZ
|
cathepsin Z
|
|
chr9_+_133320247
|
0.127
|
|
ASS1
|
argininosuccinate synthase 1
|
|
chr14_+_29236171
|
0.126
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr2_-_145090028
|
0.126
|
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr9_+_17578952
|
0.126
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr22_-_32341298
|
0.126
|
NM_015372
|
C22orf24
|
chromosome 22 open reading frame 24
|
|
chr22_+_42470247
|
0.125
|
NM_001002034
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr22_+_38864015
|
0.123
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr11_-_119293848
|
0.123
|
|
THY1
|
Thy-1 cell surface antigen
|
|
chr3_-_112358758
|
0.123
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr17_+_17206626
|
0.123
|
NM_020201
|
NT5M
|
5',3'-nucleotidase, mitochondrial
|
|
chr19_-_14048877
|
0.123
|
|
PODNL1
|
podocan-like 1
|
|
chr7_+_73082157
|
0.122
|
NM_001077621
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr7_+_73442451
|
0.122
|
|
ELN
|
elastin
|
|
chr2_-_145090038
|
0.122
|
NM_001164629
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr19_-_58609606
|
0.121
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr9_+_133320093
|
0.121
|
NM_000050
NM_054012
|
ASS1
|
argininosuccinate synthase 1
|
|
chr1_+_164528586
|
0.121
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr14_-_30396811
|
0.121
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr1_+_38273817
|
0.120
|
|
C1orf122
|
chromosome 1 open reading frame 122
|
|
chr13_-_37494370
|
0.120
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr1_+_26606621
|
0.119
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr22_+_38092994
|
0.119
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr8_-_12990731
|
0.119
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr19_-_18717467
|
0.118
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr6_+_56819923
|
0.118
|
|
BEND6
|
BEN domain containing 6
|
|
chr11_+_64781584
|
0.117
|
NM_001199745
NM_001667
|
ARL2-SNX15
ARL2
|
ARL2-SNX15 readthrough
ADP-ribosylation factor-like 2
|
|
chrX_-_134156488
|
0.117
|
NM_001078173
|
FAM127C
|
family with sequence similarity 127, member C
|
|
chr22_+_31090792
|
0.117
|
NM_030758
|
OSBP2
|
oxysterol binding protein 2
|
|
chr5_-_176739291
|
0.117
|
NM_001142935
NM_031300
|
MXD3
|
MAX dimerization protein 3
|
|
chr5_-_171710794
|
0.116
|
NM_152277
|
UBTD2
|
ubiquitin domain containing 2
|
|
chr7_+_95401817
|
0.116
|
NM_001135556
NM_001135557
NM_004411
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1
|
|
chr7_+_73442426
|
0.115
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chr20_-_31071384
|
0.115
|
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr1_-_48937791
|
0.115
|
NM_019073
|
SPATA6
|
spermatogenesis associated 6
|
|
chr1_+_26606443
|
0.115
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr11_-_2906960
|
0.114
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr7_-_131241321
|
0.113
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr19_+_14544216
|
0.113
|
|
PKN1
|
protein kinase N1
|
|
chr5_-_127873734
|
0.113
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr5_-_180688095
|
0.111
|
NM_032765
|
TRIM52
|
tripartite motif containing 52
|
|
chr16_+_12995454
|
0.111
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr9_-_130659568
|
0.111
|
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr14_-_90085325
|
0.111
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr6_+_84743250
|
0.111
|
NM_138409
|
MRAP2
|
melanocortin 2 receptor accessory protein 2
|
|
chr14_+_76618272
|
0.110
|
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chrX_+_149531439
|
0.110
|
NM_001177465
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr5_+_172483285
|
0.110
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr20_+_44462788
|
0.109
|
|
SNX21
|
sorting nexin family member 21
|
|
chr7_+_2559396
|
0.109
|
NM_001040167
NM_001040168
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr19_-_47922738
|
0.108
|
NM_001009813
NM_020160
|
MEIS3
|
Meis homeobox 3
|
|
chr20_+_34894190
|
0.108
|
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr1_+_26606586
|
0.108
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr7_-_50860665
|
0.107
|
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr19_+_14544100
|
0.107
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr16_+_89642111
|
0.106
|
NM_014427
NM_153636
|
CPNE7
|
copine VII
|
|
chr5_-_41510627
|
0.106
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr17_-_79791152
|
0.106
|
NM_001093767
NM_207368
|
FAM195B
|
family with sequence similarity 195, member B
|
|
chr8_+_104383785
|
0.106
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr15_-_48937068
|
0.106
|
|
FBN1
|
fibrillin 1
|
|
chr1_+_155829259
|
0.105
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr6_-_111804106
|
0.105
|
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr7_+_94023872
|
0.104
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr8_+_104383701
|
0.104
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr9_-_131872907
|
0.104
|
NM_000755
|
CRAT
|
carnitine O-acetyltransferase
|
|
chr4_-_775586
|
0.104
|
|
LOC100129917
|
uncharacterized LOC100129917
|
|
chr1_+_26146376
|
0.103
|
NM_001099625
NM_001099627
|
FAM54B
|
family with sequence similarity 54, member B
|
|
chr17_-_79791026
|
0.103
|
|
FAM195B
|
family with sequence similarity 195, member B
|
|
chr20_+_56964162
|
0.103
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr14_+_105941061
|
0.103
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr19_-_18654280
|
0.103
|
|
FKBP8
|
FK506 binding protein 8, 38kDa
|
|
chr20_-_2781266
|
0.103
|
NM_001184699
NM_019609
|
CPXM1
|
carboxypeptidase X (M14 family), member 1
|
|
chr1_+_26146446
|
0.103
|
|
FAM54B
|
family with sequence similarity 54, member B
|
|
chr11_+_58390145
|
0.102
|
NM_000614
|
CNTF
|
ciliary neurotrophic factor
|
|
chr9_-_37904098
|
0.102
|
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr22_+_22735202
|
0.101
|
|
|
|
|
chr7_+_66093854
|
0.101
|
NM_001167961
NM_153033
|
KCTD7
|
potassium channel tetramerisation domain containing 7
|
|
chr8_+_356933
|
0.101
|
|
FBXO25
|
F-box protein 25
|
|
chr20_+_56964289
|
0.101
|
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr6_+_3000026
|
0.100
|
NM_000904
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2
|
|
chr20_+_30946786
|
0.099
|
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr5_+_134240752
|
0.099
|
NM_032151
|
PCBD2
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2
|
|
chr7_+_28448970
|
0.098
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr20_-_2781227
|
0.098
|
|
CPXM1
|
carboxypeptidase X (M14 family), member 1
|
|
chr20_+_36531498
|
0.098
|
NM_080607
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr1_-_26232890
|
0.098
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr17_+_41177247
|
0.098
|
NM_005440
|
RND2
|
Rho family GTPase 2
|
|
chrX_-_140271275
|
0.097
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr20_+_34680569
|
0.097
|
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr11_-_2016743
|
0.097
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr9_+_139743852
|
0.096
|
|
PHPT1
|
phosphohistidine phosphatase 1
|
|
chr2_-_160919120
|
0.096
|
NM_001007267
NM_001195641
NM_007366
|
PLA2R1
|
phospholipase A2 receptor 1, 180kDa
|
|
chr11_+_45907046
|
0.096
|
NM_005456
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr5_+_134240820
|
0.096
|
|
PCBD2
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2
|
|
chr19_-_12845539
|
0.096
|
|
C19orf43
|
chromosome 19 open reading frame 43
|
|
chr1_+_948846
|
0.095
|
NM_005101
|
ISG15
|
ISG15 ubiquitin-like modifier
|
|
chr21_+_38445562
|
0.095
|
NM_001001894
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr3_-_112358510
|
0.095
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_+_139025896
|
0.094
|
NM_197964
|
C7orf55
|
chromosome 7 open reading frame 55
|
|
chr19_-_50143350
|
0.094
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr1_+_170633312
|
0.094
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr10_-_21463019
|
0.094
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr11_-_61735108
|
0.093
|
NM_002032
|
FTH1
|
ferritin, heavy polypeptide 1
|
|
chr10_-_31607919
|
0.093
|
|
ZEB1-AS1
|
ZEB1 antisense RNA 1 (non-protein coding)
|
|
chr1_+_151694012
|
0.093
|
NM_001144956
|
RIIAD1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1
|
|
chr20_-_56285576
|
0.093
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr2_+_242811880
|
0.092
|
NM_173821
|
CXXC11
|
CXXC finger protein 11
|
|
chr22_+_35777051
|
0.092
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr1_-_43232728
|
0.092
|
NM_001146289
NM_001243246
NM_022356
|
LEPRE1
|
leucine proline-enriched proteoglycan (leprecan) 1
|
|
chr5_+_40680021
|
0.092
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr3_+_187871473
|
0.092
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr19_+_35521591
|
0.092
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr1_+_26606580
|
0.091
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr19_+_15619335
|
0.091
|
NM_173483
|
CYP4F22
|
cytochrome P450, family 4, subfamily F, polypeptide 22
|
|
chr2_+_48757292
|
0.091
|
NM_001198595
NM_006873
|
STON1
|
stonin 1
|
|
chr9_+_36036900
|
0.091
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr9_+_35732207
|
0.091
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr12_+_53443834
|
0.090
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr4_-_175443601
|
0.090
|
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD)
|
|
chr20_+_34042908
|
0.089
|
NM_007186
|
CEP250
|
centrosomal protein 250kDa
|
|
chrX_-_140271293
|
0.089
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr11_-_57282315
|
0.089
|
NM_001198810
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chr10_-_79397204
|
0.089
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr8_+_81398416
|
0.089
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr12_-_54867372
|
0.089
|
NM_144594
|
GTSF1
|
gametocyte specific factor 1
|
|
chr9_-_131872782
|
0.088
|
|
CRAT
|
carnitine O-acetyltransferase
|
|
chr2_-_238322770
|
0.088
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr11_+_1889902
|
0.088
|
NM_001013253
|
LSP1
|
lymphocyte-specific protein 1
|