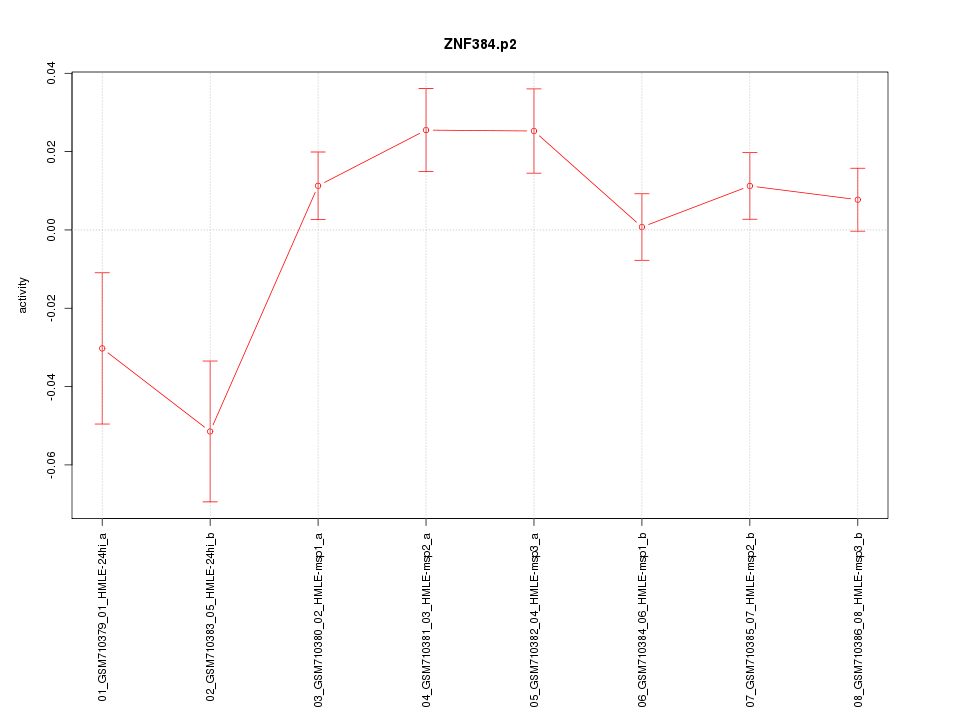
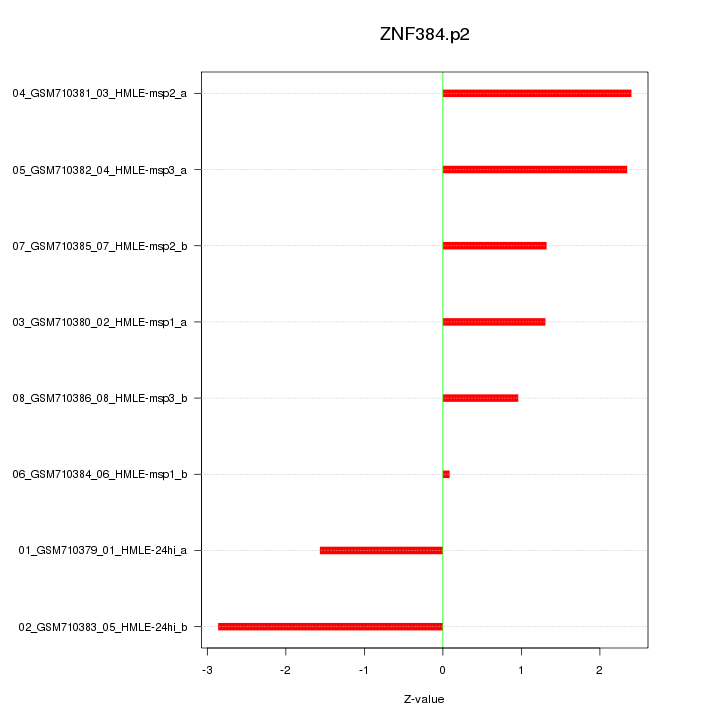
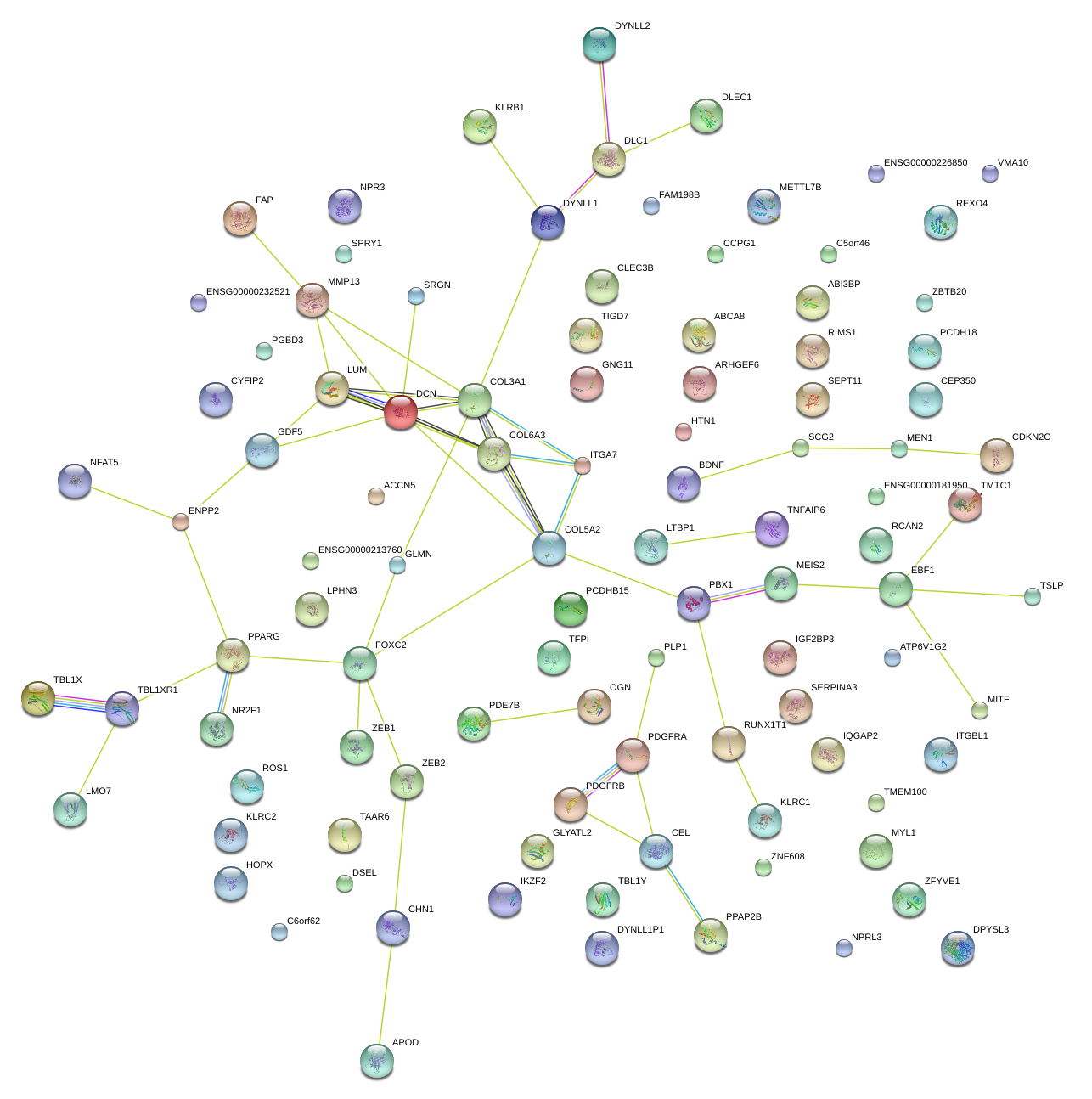
|
chr2_+_189839078
|
7.703
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189839113
|
6.867
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189839132
|
6.325
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_-_188419185
|
4.792
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr13_+_102104941
|
4.629
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr2_-_188419049
|
4.396
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr2_-_190044330
|
4.077
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr2_-_224467142
|
3.715
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr2_-_224467095
|
3.542
|
|
SCG2
|
secretogranin II
|
|
chr2_-_224467049
|
3.470
|
|
SCG2
|
secretogranin II
|
|
chr12_+_56075418
|
3.259
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr5_+_156820161
|
3.190
|
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr1_-_57045235
|
3.090
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr5_-_158526698
|
3.018
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr12_-_91576805
|
2.980
|
NM_001920
|
DCN
|
decorin
|
|
chr5_+_92918924
|
2.917
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr6_-_46293404
|
2.895
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr4_+_55095263
|
2.875
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr5_+_110407389
|
2.838
|
NM_033035
|
TSLP
|
thymic stromal lymphopoietin
|
|
chr2_+_33359607
|
2.782
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr2_-_238322770
|
2.651
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr6_+_136172802
|
2.591
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr2_-_238322840
|
2.458
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_145277879
|
2.438
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr5_-_146833150
|
2.345
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr3_+_45067750
|
2.343
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr8_-_12973752
|
2.264
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr7_-_23509631
|
2.198
|
|
|
|
|
chr2_-_238322815
|
2.101
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr10_+_70847817
|
2.030
|
NM_002727
|
SRGN
|
serglycin
|
|
chr4_-_57547845
|
1.991
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr20_-_34025955
|
1.976
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr12_-_91576579
|
1.937
|
|
DCN
|
decorin
|
|
chr4_-_156787377
|
1.917
|
NM_017419
|
ACCN5
|
amiloride-sensitive cation channel 5, intestinal
|
|
chr4_-_138453628
|
1.813
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr12_-_56106061
|
1.808
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr6_-_46293628
|
1.755
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr1_+_164528586
|
1.755
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr5_-_149535138
|
1.747
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr5_-_124080773
|
1.715
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr3_+_69985750
|
1.695
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr11_-_102826433
|
1.645
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr2_-_145277568
|
1.631
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chrX_-_135863502
|
1.602
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr12_+_56075329
|
1.577
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr5_-_158526428
|
1.577
|
|
EBF1
|
early B-cell factor 1
|
|
chr5_+_75699003
|
1.575
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr17_-_53800074
|
1.557
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr4_-_138453564
|
1.547
|
|
PCDH18
|
protocadherin 18
|
|
chrX_+_9431314
|
1.526
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr11_+_65271145
|
1.441
|
|
|
|
|
chr4_+_70916158
|
1.429
|
NM_002159
|
HTN1
|
histatin 1
|
|
chr12_-_10605928
|
1.428
|
NM_002259
NM_007328
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr17_-_66951381
|
1.424
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr5_-_149535420
|
1.421
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr4_-_159093717
|
1.419
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr8_-_120651019
|
1.406
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr9_-_95166827
|
1.401
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr4_+_124317884
|
1.367
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr16_+_69726751
|
1.357
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr3_+_12392993
|
1.327
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr15_-_37391893
|
1.303
|
|
MEIS2
|
Meis homeobox 2
|
|
chr1_-_57045214
|
1.300
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr11_-_27681195
|
1.299
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr2_-_175712276
|
1.295
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr2_-_214015053
|
1.269
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr5_+_32711437
|
1.268
|
NM_000908
NM_001204375
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr1_+_51434447
|
1.254
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr5_+_32173481
|
1.231
|
|
LOC153546
|
uncharacterized LOC153546
|
|
chr2_-_163099877
|
1.218
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr10_-_50732142
|
1.200
|
NM_170753
|
PGBD3
|
piggyBac transposable element derived 3
|
|
chr8_+_72756335
|
1.196
|
|
|
|
|
chr8_+_72756218
|
1.191
|
|
LOC100132891
|
uncharacterized LOC100132891
|
|
chr2_+_152214105
|
1.181
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr3_-_195310985
|
1.164
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr13_+_76334796
|
1.156
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr1_+_164818083
|
1.128
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr6_+_73076431
|
1.124
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr4_-_159093523
|
1.109
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr7_-_23509972
|
1.067
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr3_-_195310770
|
1.057
|
|
APOD
|
apolipoprotein D
|
|
chr4_+_62362838
|
1.033
|
NM_015236
|
LPHN3
|
latrophilin 3
|
|
chr15_-_55652777
|
1.029
|
|
CCPG1
|
cell cycle progression 1
|
|
chr4_+_124320664
|
1.025
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr7_+_93551015
|
0.998
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr18_-_65182507
|
0.996
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr5_+_140625146
|
0.995
|
NM_018935
|
PCDHB15
|
protocadherin beta 15
|
|
chr4_+_77953942
|
0.989
|
|
SEPT11
|
septin 11
|
|
chr6_-_24719286
|
0.985
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr14_+_95078713
|
0.973
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chrX_+_103031433
|
0.968
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr1_-_57045129
|
0.965
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr12_-_91505541
|
0.954
|
NM_002345
|
LUM
|
lumican
|
|
chr3_-_100712248
|
0.952
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr6_+_132891460
|
0.940
|
NM_175067
|
TAAR6
|
trace amine associated receptor 6
|
|
chr5_-_147286064
|
0.939
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr5_-_149535309
|
0.918
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr16_+_86600856
|
0.902
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr14_-_73453651
|
0.888
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr6_-_31514393
|
0.886
|
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr1_+_179965756
|
0.883
|
|
CEP350
|
centrosomal protein 350kDa
|
|
chr3_-_114790221
|
0.883
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_-_29937691
|
0.878
|
NM_175861
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr16_-_3355374
|
0.876
|
NM_033208
|
TIGD7
|
tigger transposable element derived 7
|
|
chr2_-_211179765
|
0.871
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr3_-_151987332
|
0.858
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr8_-_93111915
|
0.855
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr11_-_58611979
|
0.849
|
NM_145016
|
GLYATL2
|
glycine-N-acyltransferase-like 2
|
|
chr15_-_37391596
|
0.845
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr6_-_117747017
|
0.842
|
NM_002944
|
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase
|
|
chr14_+_101300806
|
0.841
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr3_-_112359443
|
0.832
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr22_+_38142316
|
0.826
|
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr19_-_44831907
|
0.824
|
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr1_+_162602198
|
0.813
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr1_+_164528914
|
0.809
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr15_-_37392322
|
0.807
|
|
MEIS2
|
Meis homeobox 2
|
|
chr22_+_38142255
|
0.806
|
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr16_+_4421774
|
0.805
|
NM_138440
|
VASN
|
vasorin
|
|
chr1_+_155853546
|
0.804
|
|
SYT11
|
synaptotagmin XI
|
|
chr11_+_20070609
|
0.803
|
|
NAV2
|
neuron navigator 2
|
|
chr4_-_159092509
|
0.793
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr7_-_14029286
|
0.790
|
|
ETV1
|
ets variant 1
|
|
chr3_-_112359962
|
0.779
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_162602308
|
0.773
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr7_-_14029641
|
0.772
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr11_+_65266642
|
0.771
|
|
|
|
|
chr5_-_75919051
|
0.768
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr17_+_76000248
|
0.768
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr1_-_54199876
|
0.757
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr4_-_70626429
|
0.756
|
NM_014465
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1
|
|
chr1_-_92351613
|
0.756
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr12_+_13349716
|
0.752
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr20_+_12989616
|
0.742
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr14_-_80677839
|
0.738
|
NM_001007023
NM_001242502
NM_001242503
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr1_+_201617449
|
0.730
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr1_+_183774214
|
0.729
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr11_-_2170792
|
0.726
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr10_+_12391708
|
0.701
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr1_+_51434231
|
0.699
|
NM_001262
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr9_-_73483933
|
0.691
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr5_+_67584251
|
0.687
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr14_+_101292451
|
0.685
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr16_-_89349696
|
0.684
|
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chr7_+_129932973
|
0.683
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr2_+_170366211
|
0.680
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr11_-_67141647
|
0.674
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr17_-_19651586
|
0.666
|
NM_000691
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr19_-_3029611
|
0.662
|
NM_001144762
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chrX_+_103031797
|
0.661
|
|
PLP1
|
proteolipid protein 1
|
|
chrX_+_103031753
|
0.660
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr9_+_92219926
|
0.657
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr10_-_7451198
|
0.654
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr5_+_140602914
|
0.650
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr11_-_2018703
|
0.650
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr4_+_74347461
|
0.649
|
NM_001133
|
AFM
|
afamin
|
|
chr11_-_2019070
|
0.648
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr20_-_45985411
|
0.646
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr11_-_2019007
|
0.645
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr1_-_163113938
|
0.641
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr9_+_125273080
|
0.641
|
NM_054107
|
OR1J2
|
olfactory receptor, family 1, subfamily J, member 2
|
|
chr3_+_29322802
|
0.640
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr1_+_51434375
|
0.636
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr16_+_12995454
|
0.633
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr1_-_150978879
|
0.628
|
NM_001163259
NM_001163260
NM_001040217
|
FAM63A
|
family with sequence similarity 63, member A
|
|
chr4_-_120225599
|
0.621
|
NM_001170330
|
C4orf3
|
chromosome 4 open reading frame 3
|
|
chr17_-_79791152
|
0.617
|
NM_001093767
NM_207368
|
FAM195B
|
family with sequence similarity 195, member B
|
|
chr2_-_85637486
|
0.615
|
NM_001747
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr4_-_70826712
|
0.612
|
NM_001891
|
CSN2
|
casein beta
|
|
chr1_+_174844655
|
0.607
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr5_-_111093251
|
0.606
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr16_-_73092533
|
0.606
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr4_+_55095433
|
0.601
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr1_+_196621007
|
0.600
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr5_-_39424930
|
0.594
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr1_+_201617529
|
0.592
|
|
NAV1
|
neuron navigator 1
|
|
chr21_+_47545896
|
0.589
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr1_+_104292439
|
0.588
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr5_+_140624916
|
0.588
|
|
PCDHB15
|
protocadherin beta 15
|
|
chr3_+_69812961
|
0.587
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_-_105421044
|
0.584
|
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr20_+_57470200
|
0.581
|
|
GNAS
|
GNAS complex locus
|
|
chr21_+_26934440
|
0.577
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr16_+_21689834
|
0.576
|
NM_144672
|
OTOA
|
otoancorin
|
|
chr2_-_119605758
|
0.575
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr20_+_123230
|
0.575
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr7_+_94023872
|
0.574
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chrX_+_66788682
|
0.572
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr22_+_38142230
|
0.572
|
NM_007032
NM_138632
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr13_-_44203608
|
0.570
|
NM_001127615
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr1_-_157670280
|
0.570
|
NM_052939
|
FCRL3
|
Fc receptor-like 3
|
|
chr19_-_44384240
|
0.567
|
NM_001033719
|
ZNF404
|
zinc finger protein 404
|
|
chr8_-_72756384
|
0.565
|
|
MSC
|
musculin
|
|
chr1_-_57044980
|
0.564
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr6_-_31514620
|
0.564
|
NM_001204078
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr10_+_12391471
|
0.561
|
NM_020397
NM_153498
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr17_-_63822660
|
0.555
|
NM_001037325
|
CEP112
|
centrosomal protein 112kDa
|
|
chr11_+_73358593
|
0.554
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr15_+_93443559
|
0.551
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr5_+_95066628
|
0.551
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|