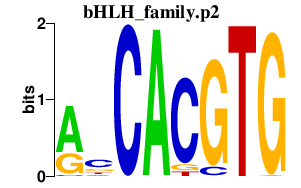
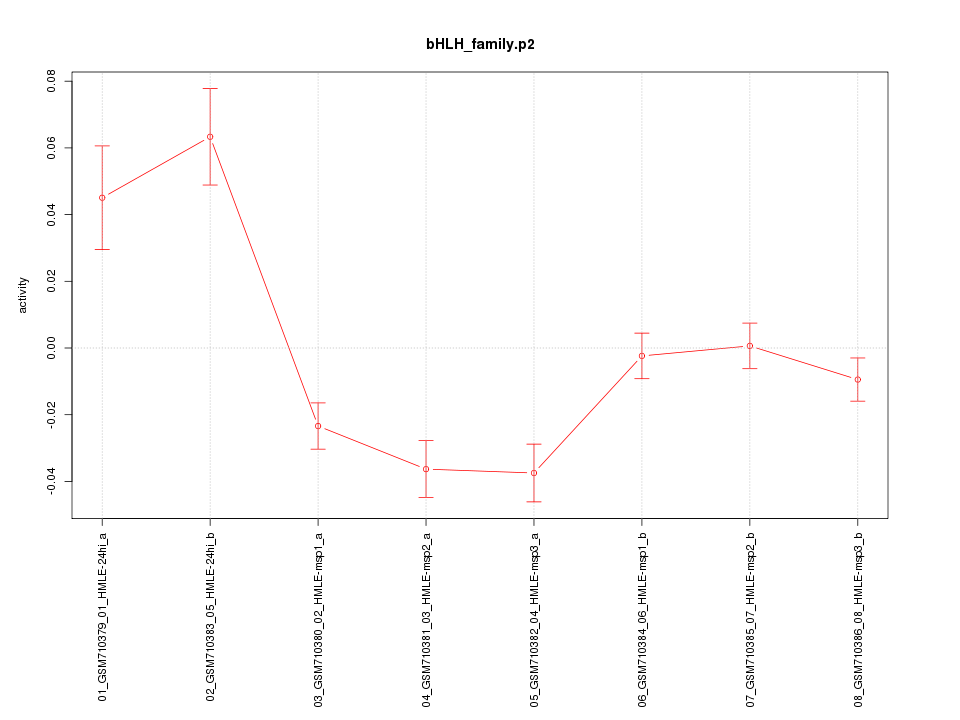
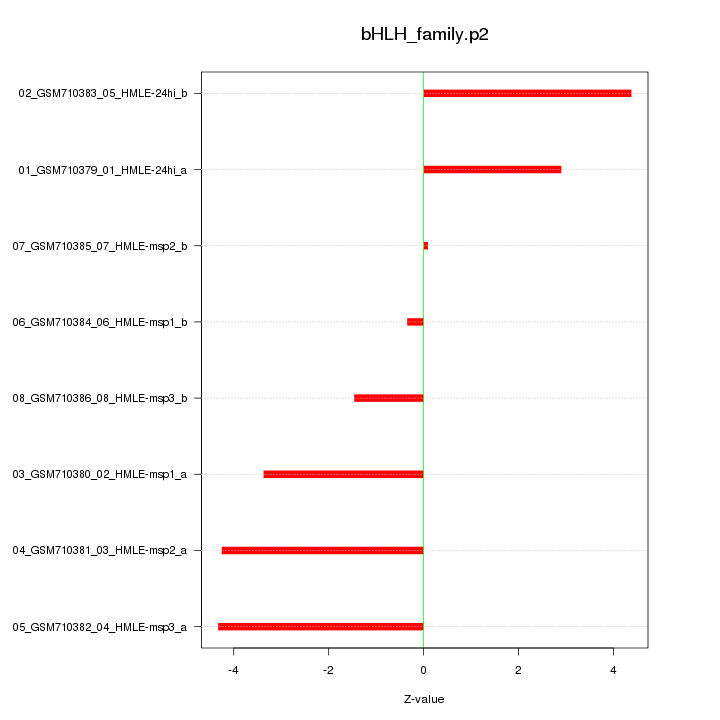
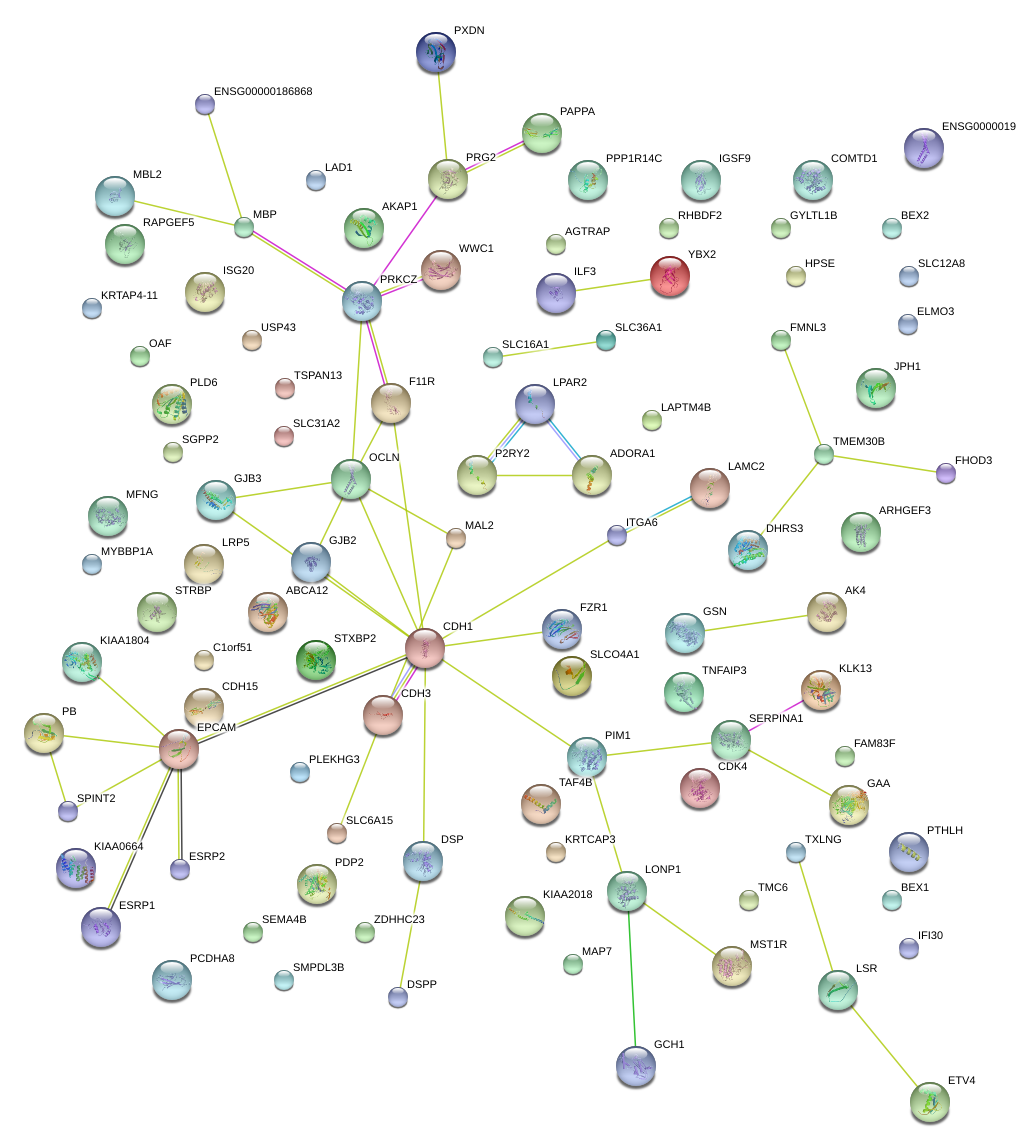
|
chr8_+_95653272
|
10.557
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr16_+_68679198
|
9.510
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr8_+_95653340
|
8.755
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr8_+_95653399
|
8.345
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr8_+_120220554
|
7.591
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr16_-_68269948
|
5.754
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr22_+_40390935
|
5.550
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr16_+_68679229
|
5.466
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr8_-_75233461
|
4.703
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr19_-_19738973
|
4.623
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr1_+_183155393
|
4.454
|
|
LAMC2
|
laminin, gamma 2
|
|
chr19_+_35739282
|
4.214
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr1_+_1981902
|
4.166
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr1_+_183155361
|
4.111
|
|
LAMC2
|
laminin, gamma 2
|
|
chr19_+_10765106
|
3.979
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr17_+_9548853
|
3.820
|
NM_153210
|
USP43
|
ubiquitin specific peptidase 43
|
|
chr7_+_16793320
|
3.807
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr14_-_61747956
|
3.598
|
|
TMEM30B
|
transmembrane protein 30B
|
|
chr6_-_136871956
|
3.413
|
NM_001198616
NM_001198617
NM_003980
|
MAP7
|
microtubule-associated protein 7
|
|
chr17_-_76124791
|
3.408
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr13_-_20767036
|
3.370
|
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr6_-_136871542
|
3.321
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr19_+_10764896
|
3.274
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr1_+_183155173
|
3.240
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr19_+_38755422
|
3.075
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr1_+_233463513
|
3.036
|
NM_032435
|
KIAA1804
|
mixed lineage kinase 4
|
|
chr1_+_150255228
|
3.030
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr3_-_57113292
|
2.925
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr11_+_72929392
|
2.911
|
NM_176071
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2
|
|
chr6_+_37137882
|
2.877
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr17_+_55163077
|
2.835
|
NM_001242903
NM_001242902
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr19_+_10764987
|
2.757
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr19_+_7702020
|
2.750
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr2_-_215896809
|
2.743
|
NM_015657
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr3_-_49941300
|
2.715
|
NM_001244937
NM_002447
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase)
|
|
chr12_-_28122884
|
2.677
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr6_+_138188352
|
2.625
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr19_+_18284578
|
2.623
|
NM_006332
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr20_+_61273823
|
2.599
|
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr2_+_27665232
|
2.560
|
NM_001168364
NM_173853
|
KRTCAP3
|
keratinocyte associated protein 3
|
|
chr19_+_7702010
|
2.539
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr19_+_38755238
|
2.519
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr17_-_17109601
|
2.500
|
|
PLD6
|
phospholipase D family, member 6
|
|
chr19_+_35739519
|
2.498
|
NM_015925
NM_205834
NM_205835
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr2_+_173292344
|
2.442
|
|
ITGA6
|
integrin, alpha 6
|
|
chr17_-_17109630
|
2.437
|
NM_178836
|
PLD6
|
phospholipase D family, member 6
|
|
chr17_+_78075390
|
2.437
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr19_+_38755097
|
2.423
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr11_+_45944155
|
2.420
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr16_+_68771192
|
2.420
|
NM_004360
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr15_+_89182038
|
2.413
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr19_+_7701976
|
2.408
|
NM_001127396
NM_006949
|
STXBP2
|
syntaxin binding protein 2
|
|
chr11_+_72929302
|
2.408
|
NM_002564
NM_176072
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2
|
|
chr17_-_4458618
|
2.405
|
NM_001105538
NM_014520
|
MYBBP1A
|
MYB binding protein (P160) 1a
|
|
chr14_-_94849552
|
2.365
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr1_-_159915382
|
2.361
|
NM_001135050
NM_020789
|
IGSF9
|
immunoglobulin superfamily, member 9
|
|
chrX_+_16804531
|
2.290
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr1_+_65613211
|
2.287
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr2_+_223289212
|
2.280
|
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr6_+_150464156
|
2.276
|
NM_030949
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C
|
|
chr18_+_23806385
|
2.272
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr5_+_140220811
|
2.254
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr16_+_68771214
|
2.251
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr11_+_68079999
|
2.227
|
NM_002335
|
LRP5
|
low density lipoprotein receptor-related protein 5
|
|
chr17_-_76124767
|
2.199
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr18_+_33877630
|
2.194
|
NM_025135
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr17_-_2614926
|
2.186
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr1_-_201368716
|
2.172
|
|
LAD1
|
ladinin 1
|
|
chr1_-_160990862
|
2.154
|
|
F11R
|
F11 receptor
|
|
chr17_-_39274587
|
2.148
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr3_-_124931562
|
2.145
|
NM_024628
|
SLC12A8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chr15_+_90728118
|
2.139
|
NM_020210
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr1_-_201368617
|
2.117
|
NM_005558
|
LAD1
|
ladinin 1
|
|
chr1_-_113498615
|
2.116
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr17_-_7197896
|
2.113
|
|
YBX2
|
Y box binding protein 2
|
|
chr5_+_68788387
|
2.104
|
NM_001205254
|
OCLN
|
occludin
|
|
chr22_-_37882214
|
2.082
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr14_+_65171243
|
2.070
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr8_+_98788014
|
2.065
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr10_-_76995685
|
2.059
|
NM_144589
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr1_-_201368434
|
2.046
|
|
LAD1
|
ladinin 1
|
|
chr18_-_74844762
|
2.045
|
NM_001025100
NM_001025101
|
MBP
|
myelin basic protein
|
|
chr9_-_126030792
|
2.027
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chrX_-_102565805
|
2.018
|
NM_001168399
NM_001168400
NM_001168401
NM_032621
|
BEX2
|
brain expressed X-linked 2
|
|
chr9_+_115913266
|
1.999
|
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr12_-_58146110
|
1.997
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr3_+_113666544
|
1.987
|
NM_173570
|
ZDHHC23
|
zinc finger, DHHC-type containing 23
|
|
chr12_-_58146025
|
1.970
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_58146108
|
1.963
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr17_-_7197866
|
1.952
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr12_-_85306539
|
1.940
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr1_+_35247838
|
1.938
|
NM_001005752
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr2_+_223289285
|
1.933
|
NM_152386
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr12_-_85306579
|
1.912
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr1_-_12677347
|
1.898
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr19_+_35739778
|
1.843
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr1_+_203097427
|
1.834
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr16_+_66914380
|
1.834
|
NM_020786
|
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2
|
|
chr9_-_126030854
|
1.807
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr12_-_58146118
|
1.804
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr1_-_160990971
|
1.783
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr17_-_7197811
|
1.781
|
|
YBX2
|
Y box binding protein 2
|
|
chr19_+_18284619
|
1.778
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr16_+_67233013
|
1.763
|
NM_024712
|
ELMO3
|
engulfment and cell motility 3
|
|
chr5_+_167718485
|
1.763
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr14_-_55369163
|
1.747
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr5_+_150827204
|
1.723
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr2_+_47596286
|
1.712
|
NM_002354
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr1_+_28261503
|
1.706
|
NM_001009568
NM_014474
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B
|
|
chr17_-_74497406
|
1.700
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr7_-_22396511
|
1.687
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr4_-_84255895
|
1.673
|
NM_001199830
|
HPSE
|
heparanase
|
|
chr1_+_11796211
|
1.666
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr19_+_18284631
|
1.650
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr17_-_41623652
|
1.649
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr4_-_84256304
|
1.640
|
NM_001166498
NM_006665
|
HPSE
|
heparanase
|
|
chr11_+_120081669
|
1.635
|
NM_178507
|
OAF
|
OAF homolog (Drosophila)
|
|
chr6_+_7541863
|
1.629
|
NM_001008844
NM_004415
|
DSP
|
desmoplakin
|
|
chr7_+_26191732
|
1.628
|
NM_004289
|
NFE2L3
|
nuclear factor (erythroid-derived 2)-like 3
|
|
chr19_+_1067293
|
1.628
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr2_-_31352489
|
1.611
|
NM_001253827
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr4_-_84255999
|
1.597
|
|
HPSE
|
heparanase
|
|
chr2_-_110873353
|
1.595
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr16_+_68771304
|
1.588
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr2_+_17721806
|
1.575
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr17_-_7165788
|
1.571
|
|
CLDN7
|
claudin 7
|
|
chr3_+_183967469
|
1.565
|
|
ECE2
|
endothelin converting enzyme 2
|
|
chr4_-_84256012
|
1.552
|
NM_001098540
|
HPSE
|
heparanase
|
|
chr9_-_140196702
|
1.546
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr2_+_17721983
|
1.545
|
|
VSNL1
|
visinin-like 1
|
|
chr2_-_208634051
|
1.540
|
NM_003468
|
FZD5
|
frizzled family receptor 5
|
|
chr11_+_1860232
|
1.534
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr6_-_80656870
|
1.521
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr12_-_85306569
|
1.521
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr16_+_2570322
|
1.518
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr8_-_15095791
|
1.507
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr1_-_113498828
|
1.506
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr3_+_183967414
|
1.502
|
NM_014693
NM_032331
|
ECE2
|
endothelin converting enzyme 2
|
|
chr1_+_11796128
|
1.501
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr2_-_110874142
|
1.494
|
NM_005434
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr11_-_61684981
|
1.491
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr6_-_143832770
|
1.485
|
NM_032020
|
FUCA2
|
fucosidase, alpha-L- 2, plasma
|
|
chr1_+_11796177
|
1.483
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr2_-_31361284
|
1.480
|
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr14_+_67999915
|
1.477
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr19_-_291335
|
1.469
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr1_+_26872306
|
1.459
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr2_-_31361591
|
1.451
|
NM_001253826
NM_024572
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr6_+_7541837
|
1.426
|
|
DSP
|
desmoplakin
|
|
chr5_-_150603615
|
1.421
|
|
CCDC69
|
coiled-coil domain containing 69
|
|
chr14_-_24036532
|
1.417
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr12_+_101673895
|
1.414
|
NM_014503
|
UTP20
|
UTP20, small subunit (SSU) processome component, homolog (yeast)
|
|
chr8_-_66701175
|
1.412
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr18_-_47721165
|
1.406
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr5_-_1882764
|
1.400
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr1_-_204329043
|
1.400
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr17_-_38657814
|
1.393
|
NM_032865
|
TNS4
|
tensin 4
|
|
chr2_-_110873638
|
1.386
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chrX_-_100662905
|
1.375
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr8_-_144654921
|
1.374
|
NM_001100878
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr11_-_11643542
|
1.373
|
NM_198516
|
GALNTL4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4
|
|
chr19_-_291168
|
1.373
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr16_+_71660063
|
1.369
|
NM_001017967
NM_052858
|
MARVELD3
|
MARVEL domain containing 3
|
|
chr11_+_7535213
|
1.365
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr13_-_20767100
|
1.361
|
NM_004004
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr1_-_21059132
|
1.360
|
NM_001103160
NM_001103161
|
SH2D5
|
SH2 domain containing 5
|
|
chr1_+_44440623
|
1.354
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr5_+_149737296
|
1.351
|
|
TCOF1
|
Treacher Collins-Franceschetti syndrome 1
|
|
chr17_-_39280377
|
1.348
|
NM_031854
|
KRTAP4-12
|
keratin associated protein 4-12
|
|
chr4_+_75310852
|
1.336
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr1_+_44440595
|
1.327
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chrX_-_100662788
|
1.323
|
|
GLA
|
galactosidase, alpha
|
|
chr3_+_184032319
|
1.310
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr12_-_121341921
|
1.310
|
|
SPPL3
|
signal peptide peptidase-like 3
|
|
chr5_+_149737201
|
1.310
|
NM_000356
NM_001008657
NM_001135243
NM_001135244
NM_001135245
NM_001195141
|
TCOF1
|
Treacher Collins-Franceschetti syndrome 1
|
|
chr4_-_103266396
|
1.305
|
NM_001135147
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr12_-_123380628
|
1.301
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr16_-_1525006
|
1.300
|
|
CLCN7
|
chloride channel 7
|
|
chr1_+_151483861
|
1.299
|
NM_020770
|
CGN
|
cingulin
|
|
chr5_+_34656367
|
1.298
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr17_-_7166263
|
1.296
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr7_+_26403944
|
1.286
|
NM_001199838
|
SNX10
|
sorting nexin 10
|
|
chr5_-_150603649
|
1.280
|
NM_015621
|
CCDC69
|
coiled-coil domain containing 69
|
|
chr19_+_1067536
|
1.278
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr5_+_34656482
|
1.275
|
|
RAI14
|
retinoic acid induced 14
|
|
chrX_+_105937067
|
1.274
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr9_-_33473846
|
1.263
|
|
NOL6
|
nucleolar protein family 6 (RNA-associated)
|
|
chr11_+_20409075
|
1.261
|
NM_001145166
NM_001145167
NM_005788
|
PRMT3
|
protein arginine methyltransferase 3
|
|
chr4_-_36245893
|
1.258
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr5_+_34656595
|
1.255
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr4_-_10023094
|
1.254
|
NM_020041
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9
|
|
chr3_+_47021146
|
1.249
|
NM_015175
|
NBEAL2
|
neurobeachin-like 2
|
|
chr6_-_13486382
|
1.249
|
NM_001242628
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr6_+_7541892
|
1.246
|
|
DSP
|
desmoplakin
|
|
chr9_-_135545327
|
1.240
|
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31
|
|
chr1_+_234349995
|
1.238
|
|
SLC35F3
|
solute carrier family 35, member F3
|
|
chr1_+_26872342
|
1.227
|
NM_001006665
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr9_+_35673852
|
1.225
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr8_+_32405727
|
1.223
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr9_+_706744
|
1.219
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|