Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for DUXA
Z-value: 0.98
Transcription factors associated with DUXA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DUXA
|
ENSG00000258873.2 | DUXA |
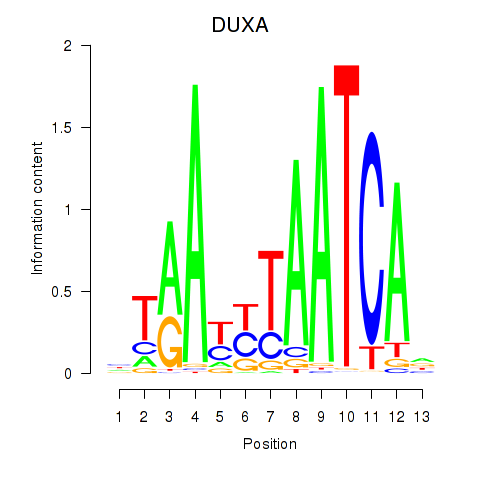
Activity profile of DUXA motif
Sorted Z-values of DUXA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DUXA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91546926 | 3.56 |
ENST00000550758.1 |
DCN |
decorin |
| chr5_-_111312622 | 1.34 |
ENST00000395634.3 |
NREP |
neuronal regeneration related protein |
| chr10_+_69865866 | 1.30 |
ENST00000354393.2 |
MYPN |
myopalladin |
| chr1_+_78956651 | 1.11 |
ENST00000370757.3 ENST00000370756.3 |
PTGFR |
prostaglandin F receptor (FP) |
| chr5_-_150537279 | 1.08 |
ENST00000517486.1 ENST00000377751.5 ENST00000356496.5 ENST00000521512.1 ENST00000517757.1 ENST00000354546.5 |
ANXA6 |
annexin A6 |
| chr1_+_78769549 | 0.86 |
ENST00000370758.1 |
PTGFR |
prostaglandin F receptor (FP) |
| chr9_-_95186739 | 0.85 |
ENST00000375550.4 |
OMD |
osteomodulin |
| chr2_-_225811747 | 0.79 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr1_+_12916941 | 0.73 |
ENST00000240189.2 |
PRAMEF2 |
PRAME family member 2 |
| chr5_-_150521192 | 0.70 |
ENST00000523714.1 ENST00000521749.1 |
ANXA6 |
annexin A6 |
| chr1_-_92351769 | 0.69 |
ENST00000212355.4 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr14_+_22615942 | 0.67 |
ENST00000390457.2 |
TRAV27 |
T cell receptor alpha variable 27 |
| chr1_-_232651312 | 0.66 |
ENST00000262861.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr1_-_72566613 | 0.64 |
ENST00000306821.3 |
NEGR1 |
neuronal growth regulator 1 |
| chr1_-_92371839 | 0.63 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr3_+_45927994 | 0.60 |
ENST00000357632.2 ENST00000395963.2 |
CCR9 |
chemokine (C-C motif) receptor 9 |
| chr2_-_202563414 | 0.58 |
ENST00000409474.3 ENST00000315506.7 ENST00000359962.5 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr16_+_21716284 | 0.57 |
ENST00000388957.3 |
OTOA |
otoancorin |
| chr5_-_75919253 | 0.52 |
ENST00000296641.4 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr8_+_70378852 | 0.51 |
ENST00000525061.1 ENST00000458141.2 ENST00000260128.4 |
SULF1 |
sulfatase 1 |
| chr1_-_13005246 | 0.48 |
ENST00000415464.2 |
PRAMEF6 |
PRAME family member 6 |
| chr3_+_69812877 | 0.48 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr10_-_49860525 | 0.46 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr1_+_78383813 | 0.46 |
ENST00000342754.5 |
NEXN |
nexilin (F actin binding protein) |
| chr3_+_69928256 | 0.45 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr11_-_96076334 | 0.44 |
ENST00000524717.1 |
MAML2 |
mastermind-like 2 (Drosophila) |
| chr1_-_12946025 | 0.43 |
ENST00000235349.5 |
PRAMEF4 |
PRAME family member 4 |
| chr13_+_76362974 | 0.43 |
ENST00000497947.2 |
LMO7 |
LIM domain 7 |
| chr5_-_75919217 | 0.42 |
ENST00000504899.1 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr1_+_13421176 | 0.41 |
ENST00000376152.1 |
PRAMEF9 |
PRAME family member 9 |
| chr4_-_142134031 | 0.40 |
ENST00000420921.2 |
RNF150 |
ring finger protein 150 |
| chr1_+_12851545 | 0.39 |
ENST00000332296.7 |
PRAMEF1 |
PRAME family member 1 |
| chr18_-_70532906 | 0.39 |
ENST00000299430.2 ENST00000397929.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr6_+_12717892 | 0.38 |
ENST00000379350.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr2_-_224467093 | 0.36 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr12_-_90049828 | 0.35 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr5_+_172571445 | 0.33 |
ENST00000231668.9 ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1 |
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr12_-_90049878 | 0.33 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr1_-_13115578 | 0.32 |
ENST00000414205.2 |
PRAMEF6 |
PRAME family member 6 |
| chr2_-_40680578 | 0.32 |
ENST00000455476.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr6_-_152639479 | 0.32 |
ENST00000356820.4 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr16_-_30006922 | 0.32 |
ENST00000564026.1 |
HIRIP3 |
HIRA interacting protein 3 |
| chr7_+_120702819 | 0.31 |
ENST00000423795.1 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chrX_+_85969626 | 0.31 |
ENST00000484479.1 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chr7_+_8008418 | 0.31 |
ENST00000223145.5 |
GLCCI1 |
glucocorticoid induced transcript 1 |
| chr12_+_26164645 | 0.31 |
ENST00000542004.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr5_+_118965244 | 0.30 |
ENST00000515256.1 ENST00000509264.1 |
FAM170A |
family with sequence similarity 170, member A |
| chr14_+_61447927 | 0.30 |
ENST00000451406.1 |
SLC38A6 |
solute carrier family 38, member 6 |
| chr14_+_61447832 | 0.29 |
ENST00000354886.2 ENST00000267488.4 |
SLC38A6 |
solute carrier family 38, member 6 |
| chr12_-_76817036 | 0.28 |
ENST00000546946.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr1_+_13359819 | 0.27 |
ENST00000376168.1 |
PRAMEF5 |
PRAME family member 5 |
| chr5_+_140571902 | 0.27 |
ENST00000239446.4 |
PCDHB10 |
protocadherin beta 10 |
| chrX_+_77166172 | 0.26 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chrX_+_54834791 | 0.26 |
ENST00000218439.4 ENST00000375058.1 ENST00000375060.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chrX_+_54835493 | 0.26 |
ENST00000396224.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr17_-_5321549 | 0.26 |
ENST00000572809.1 |
NUP88 |
nucleoporin 88kDa |
| chr1_-_165414414 | 0.26 |
ENST00000359842.5 |
RXRG |
retinoid X receptor, gamma |
| chr1_+_13641973 | 0.25 |
ENST00000330087.5 |
PRAMEF15 |
PRAME family member 15 |
| chr1_+_170501270 | 0.25 |
ENST00000367763.3 ENST00000367762.1 |
GORAB |
golgin, RAB6-interacting |
| chr2_+_228678550 | 0.25 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chr5_+_140723601 | 0.24 |
ENST00000253812.6 |
PCDHGA3 |
protocadherin gamma subfamily A, 3 |
| chr2_+_109223595 | 0.24 |
ENST00000410093.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr3_+_108321623 | 0.24 |
ENST00000497905.1 ENST00000463306.1 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
| chr8_-_86575726 | 0.24 |
ENST00000379010.2 |
REXO1L1 |
REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 1, pseudogene |
| chr17_+_66511540 | 0.24 |
ENST00000588188.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr3_-_164914640 | 0.24 |
ENST00000241274.3 |
SLITRK3 |
SLIT and NTRK-like family, member 3 |
| chr14_-_107114267 | 0.23 |
ENST00000454421.2 |
IGHV3-64 |
immunoglobulin heavy variable 3-64 |
| chr19_-_3061397 | 0.23 |
ENST00000586839.1 |
AES |
amino-terminal enhancer of split |
| chr16_+_77756399 | 0.22 |
ENST00000564085.1 ENST00000268533.5 ENST00000568787.1 ENST00000437314.3 ENST00000563839.1 |
NUDT7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr2_+_161993465 | 0.22 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr5_+_154238149 | 0.22 |
ENST00000519430.1 ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr17_-_7145475 | 0.21 |
ENST00000571129.1 ENST00000571253.1 ENST00000573928.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr2_+_201994042 | 0.21 |
ENST00000417748.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr12_+_26126681 | 0.21 |
ENST00000542865.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr6_+_52285046 | 0.21 |
ENST00000371068.5 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
| chr6_+_28092338 | 0.21 |
ENST00000340487.4 |
ZSCAN16 |
zinc finger and SCAN domain containing 16 |
| chr7_-_137686791 | 0.21 |
ENST00000452463.1 ENST00000330387.6 ENST00000456390.1 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
| chr6_+_73076432 | 0.21 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr20_-_44298878 | 0.20 |
ENST00000324384.3 ENST00000356562.2 |
WFDC11 |
WAP four-disulfide core domain 11 |
| chr14_+_53019993 | 0.20 |
ENST00000542169.2 ENST00000555622.1 |
GPR137C |
G protein-coupled receptor 137C |
| chr2_-_198062758 | 0.20 |
ENST00000328737.2 |
ANKRD44 |
ankyrin repeat domain 44 |
| chr4_-_119759795 | 0.20 |
ENST00000419654.2 |
SEC24D |
SEC24 family member D |
| chr5_+_140079919 | 0.20 |
ENST00000274712.3 |
ZMAT2 |
zinc finger, matrin-type 2 |
| chr1_-_27998689 | 0.19 |
ENST00000339145.4 ENST00000362020.4 ENST00000361157.6 |
IFI6 |
interferon, alpha-inducible protein 6 |
| chr1_+_104159999 | 0.19 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr6_+_167704838 | 0.19 |
ENST00000366829.2 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr6_+_167704798 | 0.19 |
ENST00000230256.3 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr12_-_81763184 | 0.19 |
ENST00000548670.1 ENST00000541570.2 ENST00000553058.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr8_+_58890917 | 0.19 |
ENST00000522992.1 |
RP11-1112C15.1 |
RP11-1112C15.1 |
| chr4_-_23735183 | 0.19 |
ENST00000507666.1 |
RP11-380P13.2 |
RP11-380P13.2 |
| chr5_+_98109322 | 0.19 |
ENST00000513185.1 |
RGMB |
repulsive guidance molecule family member b |
| chr1_-_13007420 | 0.19 |
ENST00000376189.1 |
PRAMEF6 |
PRAME family member 6 |
| chr12_+_8662057 | 0.18 |
ENST00000382064.2 |
CLEC4D |
C-type lectin domain family 4, member D |
| chr12_-_81763127 | 0.18 |
ENST00000541017.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr19_-_6393216 | 0.18 |
ENST00000595047.1 |
GTF2F1 |
general transcription factor IIF, polypeptide 1, 74kDa |
| chr3_-_195997410 | 0.18 |
ENST00000419333.1 |
PCYT1A |
phosphate cytidylyltransferase 1, choline, alpha |
| chr5_+_154237778 | 0.18 |
ENST00000523698.1 ENST00000517876.1 ENST00000520472.1 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr5_+_154238096 | 0.18 |
ENST00000517568.1 ENST00000524105.1 ENST00000285896.6 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr5_+_154238042 | 0.18 |
ENST00000519211.1 ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr2_+_161993412 | 0.17 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr7_+_134331550 | 0.16 |
ENST00000344924.3 ENST00000418040.1 ENST00000393132.2 |
BPGM |
2,3-bisphosphoglycerate mutase |
| chr9_+_139877445 | 0.16 |
ENST00000408973.2 |
LCNL1 |
lipocalin-like 1 |
| chr17_-_7145106 | 0.16 |
ENST00000577035.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr12_-_110434183 | 0.16 |
ENST00000360185.4 ENST00000354574.4 ENST00000338373.5 ENST00000343646.5 ENST00000356259.4 ENST00000553118.1 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr19_-_12833361 | 0.15 |
ENST00000592287.1 |
TNPO2 |
transportin 2 |
| chr12_-_10607084 | 0.15 |
ENST00000408006.3 ENST00000544822.1 ENST00000536188.1 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr10_+_82297658 | 0.15 |
ENST00000339284.2 |
SH2D4B |
SH2 domain containing 4B |
| chr1_+_95616933 | 0.15 |
ENST00000604203.1 |
RP11-57H12.6 |
TMEM56-RWDD3 readthrough |
| chr2_-_131099897 | 0.15 |
ENST00000409127.1 ENST00000437688.2 ENST00000259229.2 |
CCDC115 |
coiled-coil domain containing 115 |
| chr5_+_81601166 | 0.15 |
ENST00000439350.1 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chrX_+_100878079 | 0.15 |
ENST00000471229.2 |
ARMCX3 |
armadillo repeat containing, X-linked 3 |
| chr15_+_44719970 | 0.15 |
ENST00000558966.1 |
CTDSPL2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr5_-_140700322 | 0.14 |
ENST00000313368.5 |
TAF7 |
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr14_-_75536182 | 0.14 |
ENST00000555463.1 |
ACYP1 |
acylphosphatase 1, erythrocyte (common) type |
| chr12_-_110434021 | 0.14 |
ENST00000355312.3 ENST00000551209.1 ENST00000550186.1 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr13_+_20207782 | 0.14 |
ENST00000414242.2 ENST00000361479.5 |
MPHOSPH8 |
M-phase phosphoprotein 8 |
| chr6_+_31126291 | 0.14 |
ENST00000376257.3 ENST00000376255.4 |
TCF19 |
transcription factor 19 |
| chr1_-_12908578 | 0.14 |
ENST00000317869.6 |
HNRNPCL1 |
heterogeneous nuclear ribonucleoprotein C-like 1 |
| chr2_+_101437487 | 0.13 |
ENST00000427413.1 ENST00000542504.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr8_-_6914251 | 0.13 |
ENST00000330590.2 |
DEFA5 |
defensin, alpha 5, Paneth cell-specific |
| chr20_-_33999766 | 0.13 |
ENST00000349714.5 ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr17_-_39553844 | 0.13 |
ENST00000251645.2 |
KRT31 |
keratin 31 |
| chr2_+_169923577 | 0.13 |
ENST00000432060.2 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr2_+_54342574 | 0.12 |
ENST00000303536.4 ENST00000394666.3 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr19_-_17488143 | 0.12 |
ENST00000599426.1 ENST00000252590.4 |
PLVAP |
plasmalemma vesicle associated protein |
| chr1_+_53480598 | 0.12 |
ENST00000430330.2 ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2 |
sterol carrier protein 2 |
| chr12_-_102874102 | 0.12 |
ENST00000392905.2 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr8_-_7673238 | 0.12 |
ENST00000335021.2 |
DEFB107A |
defensin, beta 107A |
| chr15_-_65407524 | 0.12 |
ENST00000559089.1 |
UBAP1L |
ubiquitin associated protein 1-like |
| chr12_-_54653313 | 0.12 |
ENST00000550411.1 ENST00000439541.2 |
CBX5 |
chromobox homolog 5 |
| chr3_-_47950745 | 0.12 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr9_-_6015607 | 0.12 |
ENST00000259569.5 |
RANBP6 |
RAN binding protein 6 |
| chr1_-_19615744 | 0.12 |
ENST00000361640.4 |
AKR7A3 |
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr16_+_30006997 | 0.12 |
ENST00000304516.7 |
INO80E |
INO80 complex subunit E |
| chr5_+_54320078 | 0.12 |
ENST00000231009.2 |
GZMK |
granzyme K (granzyme 3; tryptase II) |
| chr16_-_57219966 | 0.11 |
ENST00000565760.1 ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A |
family with sequence similarity 192, member A |
| chr2_-_10587897 | 0.11 |
ENST00000405333.1 ENST00000443218.1 |
ODC1 |
ornithine decarboxylase 1 |
| chr16_+_69221028 | 0.11 |
ENST00000336278.4 |
SNTB2 |
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr7_-_137028498 | 0.11 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr1_-_13117736 | 0.11 |
ENST00000376192.5 ENST00000376182.1 |
PRAMEF6 |
PRAME family member 6 |
| chr15_+_39542867 | 0.11 |
ENST00000318578.3 ENST00000561223.1 |
C15orf54 |
chromosome 15 open reading frame 54 |
| chr15_+_71228826 | 0.11 |
ENST00000558456.1 ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr3_+_186353756 | 0.11 |
ENST00000431018.1 ENST00000450521.1 ENST00000539949.1 |
FETUB |
fetuin B |
| chr2_+_201994208 | 0.11 |
ENST00000440180.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr3_+_159943362 | 0.11 |
ENST00000326474.3 |
C3orf80 |
chromosome 3 open reading frame 80 |
| chr1_+_206557366 | 0.11 |
ENST00000414007.1 ENST00000419187.2 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
| chr8_+_7353368 | 0.11 |
ENST00000355602.2 |
DEFB107B |
defensin, beta 107B |
| chr5_+_149877334 | 0.11 |
ENST00000523767.1 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr19_-_47349395 | 0.11 |
ENST00000597020.1 |
AP2S1 |
adaptor-related protein complex 2, sigma 1 subunit |
| chr10_+_134150835 | 0.11 |
ENST00000432555.2 |
LRRC27 |
leucine rich repeat containing 27 |
| chr1_-_12958101 | 0.11 |
ENST00000235347.4 |
PRAMEF10 |
PRAME family member 10 |
| chr2_+_169923504 | 0.11 |
ENST00000357546.2 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr17_+_66521936 | 0.10 |
ENST00000592800.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr17_-_79849438 | 0.10 |
ENST00000331204.4 ENST00000505490.2 |
ALYREF |
Aly/REF export factor |
| chr10_+_180643 | 0.10 |
ENST00000509513.2 ENST00000397959.3 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
| chr16_+_30007524 | 0.10 |
ENST00000567254.1 ENST00000567705.1 |
INO80E |
INO80 complex subunit E |
| chr7_-_137028534 | 0.10 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chr19_+_507299 | 0.10 |
ENST00000359315.5 |
TPGS1 |
tubulin polyglutamylase complex subunit 1 |
| chr12_+_72332618 | 0.10 |
ENST00000333850.3 |
TPH2 |
tryptophan hydroxylase 2 |
| chr4_+_148653206 | 0.10 |
ENST00000336498.3 |
ARHGAP10 |
Rho GTPase activating protein 10 |
| chr9_+_140445651 | 0.10 |
ENST00000371443.5 |
MRPL41 |
mitochondrial ribosomal protein L41 |
| chr11_+_62496114 | 0.10 |
ENST00000532583.1 |
TTC9C |
tetratricopeptide repeat domain 9C |
| chr16_-_89785777 | 0.10 |
ENST00000561976.1 |
VPS9D1 |
VPS9 domain containing 1 |
| chr11_-_6462210 | 0.10 |
ENST00000265983.3 |
HPX |
hemopexin |
| chr16_-_32688053 | 0.10 |
ENST00000398682.4 |
TP53TG3 |
TP53 target 3 |
| chr2_+_201994569 | 0.10 |
ENST00000457277.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr14_-_80697396 | 0.10 |
ENST00000557010.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr16_+_32264040 | 0.10 |
ENST00000398664.3 |
TP53TG3D |
TP53 target 3D |
| chr5_+_68860949 | 0.10 |
ENST00000507595.1 |
GTF2H2C |
general transcription factor IIH, polypeptide 2C |
| chr6_+_35996859 | 0.09 |
ENST00000472333.1 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr5_+_102201687 | 0.09 |
ENST00000304400.7 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr12_+_12878829 | 0.09 |
ENST00000326765.6 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr6_+_116691001 | 0.09 |
ENST00000537543.1 |
DSE |
dermatan sulfate epimerase |
| chr2_+_54342533 | 0.09 |
ENST00000406041.1 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr3_-_149093499 | 0.09 |
ENST00000472441.1 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr1_-_157811588 | 0.09 |
ENST00000368174.4 |
CD5L |
CD5 molecule-like |
| chr5_+_102201722 | 0.09 |
ENST00000274392.9 ENST00000455264.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr17_-_4167142 | 0.09 |
ENST00000570535.1 ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
| chrX_+_46306624 | 0.09 |
ENST00000360017.5 |
KRBOX4 |
KRAB box domain containing 4 |
| chr4_-_69215699 | 0.09 |
ENST00000510746.1 ENST00000344157.4 ENST00000355665.3 |
YTHDC1 |
YTH domain containing 1 |
| chr18_+_3449330 | 0.09 |
ENST00000549253.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr14_+_55595762 | 0.09 |
ENST00000254301.9 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr11_-_18610246 | 0.08 |
ENST00000379387.4 ENST00000541984.1 |
UEVLD |
UEV and lactate/malate dehyrogenase domains |
| chr5_-_16742330 | 0.08 |
ENST00000505695.1 ENST00000427430.2 |
MYO10 |
myosin X |
| chr8_-_62602327 | 0.08 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr2_-_161350305 | 0.08 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr7_+_134832808 | 0.08 |
ENST00000275767.3 |
TMEM140 |
transmembrane protein 140 |
| chr8_+_24151620 | 0.08 |
ENST00000437154.2 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr11_+_67250490 | 0.08 |
ENST00000528641.2 ENST00000279146.3 |
AIP |
aryl hydrocarbon receptor interacting protein |
| chr10_+_18689637 | 0.08 |
ENST00000377315.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr20_-_25371412 | 0.08 |
ENST00000339157.5 ENST00000376542.3 |
ABHD12 |
abhydrolase domain containing 12 |
| chr7_+_23338819 | 0.08 |
ENST00000466681.1 |
MALSU1 |
mitochondrial assembly of ribosomal large subunit 1 |
| chr11_+_43333513 | 0.08 |
ENST00000534695.1 ENST00000455725.2 ENST00000531273.1 ENST00000420461.2 ENST00000378852.3 ENST00000534600.1 |
API5 |
apoptosis inhibitor 5 |
| chr4_+_169552748 | 0.08 |
ENST00000504519.1 ENST00000512127.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr5_+_54455946 | 0.08 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chr20_+_22034809 | 0.08 |
ENST00000449427.1 |
RP11-125P18.1 |
RP11-125P18.1 |
| chr6_-_133055896 | 0.07 |
ENST00000367927.5 ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3 |
vanin 3 |
| chr4_+_646960 | 0.07 |
ENST00000488061.1 ENST00000429163.2 |
PDE6B |
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chrX_-_154689596 | 0.07 |
ENST00000369444.2 |
H2AFB3 |
H2A histone family, member B3 |
| chr6_-_31125850 | 0.07 |
ENST00000507751.1 ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1 |
coiled-coil alpha-helical rod protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) response to luteinizing hormone(GO:0034699) |
| 0.2 | 3.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 2.0 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.3 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 0.7 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.2 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.8 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.6 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.8 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.4 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.9 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.9 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.6 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.3 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.2 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.0 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0015014 | protein sulfation(GO:0006477) heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 1.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.0 | GO:0090264 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of eosinophil degranulation(GO:0043309) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 3.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.9 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 1.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.2 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 1.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 3.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 1.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 3.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |