Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
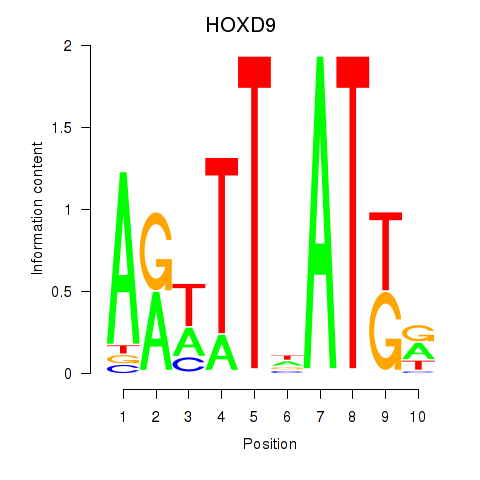
Results for HOXD9
Z-value: 0.56
Transcription factors associated with HOXD9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD9
|
ENSG00000128709.10 | HOXD9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD9 | hg19_v2_chr2_+_176987088_176987088 | 0.79 | 1.9e-02 | Click! |
Activity profile of HOXD9 motif
Sorted Z-values of HOXD9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_61554932 | 0.62 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr18_+_21452804 | 0.60 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr11_-_104972158 | 0.49 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr10_-_105845674 | 0.49 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chrX_-_33229636 | 0.33 |
ENST00000357033.4 |
DMD |
dystrophin |
| chr11_+_69924639 | 0.32 |
ENST00000538023.1 ENST00000398543.2 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr5_+_147582348 | 0.31 |
ENST00000514389.1 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr1_+_152956549 | 0.30 |
ENST00000307122.2 |
SPRR1A |
small proline-rich protein 1A |
| chr7_+_16793160 | 0.27 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr1_+_81771806 | 0.23 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr11_-_104905840 | 0.23 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chr6_+_132891461 | 0.21 |
ENST00000275198.1 |
TAAR6 |
trace amine associated receptor 6 |
| chr8_+_104892639 | 0.21 |
ENST00000436393.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr19_+_45843994 | 0.18 |
ENST00000391946.2 |
KLC3 |
kinesin light chain 3 |
| chr4_-_84035868 | 0.17 |
ENST00000426923.2 ENST00000509973.1 |
PLAC8 |
placenta-specific 8 |
| chr11_+_123986069 | 0.17 |
ENST00000456829.2 ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A |
von Willebrand factor A domain containing 5A |
| chr3_+_145782358 | 0.16 |
ENST00000422482.1 |
AC107021.1 |
HCG1786590; PRO2533; Uncharacterized protein |
| chr5_+_140180635 | 0.16 |
ENST00000522353.2 ENST00000532566.2 |
PCDHA3 |
protocadherin alpha 3 |
| chr4_-_84035905 | 0.16 |
ENST00000311507.4 |
PLAC8 |
placenta-specific 8 |
| chr5_+_140248518 | 0.16 |
ENST00000398640.2 |
PCDHA11 |
protocadherin alpha 11 |
| chr4_+_69313145 | 0.15 |
ENST00000305363.4 |
TMPRSS11E |
transmembrane protease, serine 11E |
| chr6_+_32812568 | 0.15 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr8_+_86089460 | 0.15 |
ENST00000418930.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr4_-_76944621 | 0.14 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr10_+_60759378 | 0.14 |
ENST00000432535.1 |
LINC00844 |
long intergenic non-protein coding RNA 844 |
| chr4_-_110723194 | 0.14 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr8_+_86089619 | 0.14 |
ENST00000256117.5 ENST00000416274.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr5_+_147582387 | 0.14 |
ENST00000325630.2 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr1_+_160370344 | 0.14 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr17_-_39646116 | 0.13 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr5_+_140207536 | 0.13 |
ENST00000529310.1 ENST00000527624.1 |
PCDHA6 |
protocadherin alpha 6 |
| chr7_-_107880508 | 0.13 |
ENST00000425651.2 |
NRCAM |
neuronal cell adhesion molecule |
| chr12_+_26348246 | 0.12 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr3_-_139258521 | 0.12 |
ENST00000483943.2 ENST00000232219.2 ENST00000492918.1 |
RBP1 |
retinol binding protein 1, cellular |
| chr18_-_53069419 | 0.12 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr1_+_16083154 | 0.12 |
ENST00000375771.1 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr8_+_52730143 | 0.11 |
ENST00000415643.1 |
AC090186.1 |
Uncharacterized protein |
| chr8_-_86253888 | 0.11 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr6_+_1312675 | 0.11 |
ENST00000296839.2 |
FOXQ1 |
forkhead box Q1 |
| chr12_-_95510743 | 0.11 |
ENST00000551521.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr1_-_59043166 | 0.11 |
ENST00000371225.2 |
TACSTD2 |
tumor-associated calcium signal transducer 2 |
| chr6_+_72926145 | 0.11 |
ENST00000425662.2 ENST00000453976.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr3_-_116163830 | 0.11 |
ENST00000333617.4 |
LSAMP |
limbic system-associated membrane protein |
| chr14_+_23654525 | 0.11 |
ENST00000399910.1 ENST00000492621.1 |
C14orf164 |
chromosome 14 open reading frame 164 |
| chr6_+_73076432 | 0.10 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr8_-_66701319 | 0.10 |
ENST00000379419.4 |
PDE7A |
phosphodiesterase 7A |
| chr12_+_19358228 | 0.10 |
ENST00000424268.1 ENST00000543806.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr7_-_111032971 | 0.10 |
ENST00000450877.1 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr11_-_13517565 | 0.10 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr11_-_16430399 | 0.09 |
ENST00000528252.1 |
SOX6 |
SRY (sex determining region Y)-box 6 |
| chr5_-_78808617 | 0.09 |
ENST00000282260.6 ENST00000508576.1 ENST00000535690.1 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr4_+_175839551 | 0.09 |
ENST00000404450.4 ENST00000514159.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr12_-_88974236 | 0.09 |
ENST00000228280.5 ENST00000552044.1 ENST00000357116.4 |
KITLG |
KIT ligand |
| chr2_+_54342574 | 0.09 |
ENST00000303536.4 ENST00000394666.3 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr16_-_20556492 | 0.09 |
ENST00000568098.1 |
ACSM2B |
acyl-CoA synthetase medium-chain family member 2B |
| chr11_-_104827425 | 0.08 |
ENST00000393150.3 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr12_+_119772502 | 0.08 |
ENST00000536742.1 ENST00000327554.2 |
CCDC60 |
coiled-coil domain containing 60 |
| chr2_+_185463093 | 0.08 |
ENST00000302277.6 |
ZNF804A |
zinc finger protein 804A |
| chr2_+_145780725 | 0.08 |
ENST00000451478.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr2_+_145780767 | 0.08 |
ENST00000599358.1 ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr6_+_13272904 | 0.08 |
ENST00000379335.3 ENST00000379329.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr13_-_44735393 | 0.07 |
ENST00000400419.1 |
SMIM2 |
small integral membrane protein 2 |
| chr9_+_12693336 | 0.07 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr11_-_102401469 | 0.07 |
ENST00000260227.4 |
MMP7 |
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr1_+_45140360 | 0.07 |
ENST00000418644.1 ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228 |
chromosome 1 open reading frame 228 |
| chr1_+_196857144 | 0.07 |
ENST00000367416.2 ENST00000251424.4 ENST00000367418.2 |
CFHR4 |
complement factor H-related 4 |
| chr2_+_145780739 | 0.07 |
ENST00000597173.1 ENST00000602108.1 ENST00000420472.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr11_-_5255861 | 0.07 |
ENST00000380299.3 |
HBD |
hemoglobin, delta |
| chr6_-_52774464 | 0.07 |
ENST00000370968.1 ENST00000211122.3 |
GSTA3 |
glutathione S-transferase alpha 3 |
| chr6_-_132967142 | 0.06 |
ENST00000275216.1 |
TAAR1 |
trace amine associated receptor 1 |
| chrX_-_130423200 | 0.06 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr3_+_97887544 | 0.06 |
ENST00000356526.2 |
OR5H15 |
olfactory receptor, family 5, subfamily H, member 15 |
| chr1_+_101185290 | 0.06 |
ENST00000370119.4 ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1 |
vascular cell adhesion molecule 1 |
| chr6_-_32634425 | 0.06 |
ENST00000399082.3 ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1 |
major histocompatibility complex, class II, DQ beta 1 |
| chr17_+_68071389 | 0.06 |
ENST00000283936.1 ENST00000392671.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr18_+_616672 | 0.06 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr12_-_8765446 | 0.06 |
ENST00000537228.1 ENST00000229335.6 |
AICDA |
activation-induced cytidine deaminase |
| chr11_-_105010320 | 0.06 |
ENST00000532895.1 ENST00000530950.1 |
CARD18 |
caspase recruitment domain family, member 18 |
| chr19_-_5903714 | 0.06 |
ENST00000586349.1 ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12 NDUFA11 FUT5 |
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chrX_+_144908928 | 0.06 |
ENST00000408967.2 |
TMEM257 |
transmembrane protein 257 |
| chr17_-_10452929 | 0.06 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr6_+_18387570 | 0.06 |
ENST00000259939.3 |
RNF144B |
ring finger protein 144B |
| chr3_+_182511266 | 0.06 |
ENST00000323116.5 ENST00000493826.1 |
ATP11B |
ATPase, class VI, type 11B |
| chr3_+_94657086 | 0.05 |
ENST00000463200.1 |
LINC00879 |
long intergenic non-protein coding RNA 879 |
| chr1_+_38022513 | 0.05 |
ENST00000296218.7 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr10_-_49459800 | 0.05 |
ENST00000305531.3 |
FRMPD2 |
FERM and PDZ domain containing 2 |
| chr4_+_25915822 | 0.05 |
ENST00000506197.2 |
SMIM20 |
small integral membrane protein 20 |
| chr2_+_143635067 | 0.05 |
ENST00000264170.4 |
KYNU |
kynureninase |
| chr17_+_68071458 | 0.05 |
ENST00000589377.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_-_10607084 | 0.05 |
ENST00000408006.3 ENST00000544822.1 ENST00000536188.1 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr4_-_69434245 | 0.05 |
ENST00000317746.2 |
UGT2B17 |
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr3_+_158787041 | 0.05 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr22_+_30477000 | 0.05 |
ENST00000403975.1 |
HORMAD2 |
HORMA domain containing 2 |
| chr1_+_87012753 | 0.05 |
ENST00000370563.3 |
CLCA4 |
chloride channel accessory 4 |
| chr10_-_13523073 | 0.05 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr19_+_55014085 | 0.05 |
ENST00000351841.2 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
| chr12_-_110486348 | 0.05 |
ENST00000547573.1 ENST00000546651.2 ENST00000551185.2 |
C12orf76 |
chromosome 12 open reading frame 76 |
| chr3_+_140981456 | 0.05 |
ENST00000504264.1 |
ACPL2 |
acid phosphatase-like 2 |
| chr22_+_50609150 | 0.05 |
ENST00000159647.5 ENST00000395842.2 |
PANX2 |
pannexin 2 |
| chr11_+_112047087 | 0.05 |
ENST00000526088.1 ENST00000532593.1 ENST00000531169.1 |
BCO2 |
beta-carotene oxygenase 2 |
| chr7_-_121944491 | 0.05 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr8_-_102181718 | 0.05 |
ENST00000565617.1 |
KB-1460A1.5 |
KB-1460A1.5 |
| chr14_+_39583427 | 0.05 |
ENST00000308317.6 ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2 |
gem (nuclear organelle) associated protein 2 |
| chr19_+_55014013 | 0.05 |
ENST00000301202.2 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
| chr8_+_107738343 | 0.05 |
ENST00000521592.1 |
OXR1 |
oxidation resistance 1 |
| chr3_+_94657118 | 0.04 |
ENST00000466089.1 ENST00000470465.1 |
LINC00879 |
long intergenic non-protein coding RNA 879 |
| chrX_-_72434628 | 0.04 |
ENST00000536638.1 ENST00000373517.3 |
NAP1L2 |
nucleosome assembly protein 1-like 2 |
| chr3_+_158991025 | 0.04 |
ENST00000337808.6 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr19_+_10397648 | 0.04 |
ENST00000340992.4 ENST00000393717.2 |
ICAM4 |
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr18_+_616711 | 0.04 |
ENST00000579494.1 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr13_-_47012325 | 0.04 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr2_-_118943930 | 0.04 |
ENST00000449075.1 ENST00000414886.1 ENST00000449819.1 |
AC093901.1 |
AC093901.1 |
| chr9_-_71629010 | 0.04 |
ENST00000377276.2 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
| chr21_+_17792672 | 0.04 |
ENST00000602620.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr1_+_104615595 | 0.04 |
ENST00000418362.1 |
RP11-364B6.1 |
RP11-364B6.1 |
| chr1_-_39339777 | 0.04 |
ENST00000397572.2 |
MYCBP |
MYC binding protein |
| chr15_-_90222642 | 0.04 |
ENST00000430628.2 |
PLIN1 |
perilipin 1 |
| chr4_-_72649763 | 0.04 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr15_+_57998923 | 0.04 |
ENST00000380557.4 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
| chr15_-_90222610 | 0.04 |
ENST00000300055.5 |
PLIN1 |
perilipin 1 |
| chrX_-_125686784 | 0.04 |
ENST00000371126.1 |
DCAF12L1 |
DDB1 and CUL4 associated factor 12-like 1 |
| chrX_-_68385274 | 0.04 |
ENST00000374584.3 ENST00000590146.1 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
| chr9_-_39239171 | 0.04 |
ENST00000358144.2 |
CNTNAP3 |
contactin associated protein-like 3 |
| chr3_-_99569821 | 0.04 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr7_-_50633078 | 0.04 |
ENST00000444124.2 |
DDC |
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr6_-_88411911 | 0.04 |
ENST00000257787.5 |
AKIRIN2 |
akirin 2 |
| chr15_-_66797172 | 0.04 |
ENST00000569438.1 ENST00000569696.1 ENST00000307961.6 |
RPL4 |
ribosomal protein L4 |
| chr2_+_210517895 | 0.04 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr2_+_1418154 | 0.04 |
ENST00000423320.1 ENST00000382198.1 |
TPO |
thyroid peroxidase |
| chr8_-_10512569 | 0.04 |
ENST00000382483.3 |
RP1L1 |
retinitis pigmentosa 1-like 1 |
| chr11_+_63606373 | 0.04 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr17_+_61086917 | 0.04 |
ENST00000424789.2 ENST00000389520.4 |
TANC2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chrX_-_68385354 | 0.04 |
ENST00000361478.1 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
| chr17_+_9479944 | 0.03 |
ENST00000396219.3 ENST00000352665.5 |
WDR16 |
WD repeat domain 16 |
| chr2_+_54342533 | 0.03 |
ENST00000406041.1 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr12_-_9268707 | 0.03 |
ENST00000318602.7 |
A2M |
alpha-2-macroglobulin |
| chr7_-_16921601 | 0.03 |
ENST00000402239.3 ENST00000310398.2 ENST00000414935.1 |
AGR3 |
anterior gradient 3 |
| chr2_+_135809835 | 0.03 |
ENST00000264158.8 ENST00000539493.1 ENST00000442034.1 ENST00000425393.1 |
RAB3GAP1 |
RAB3 GTPase activating protein subunit 1 (catalytic) |
| chr16_+_4545853 | 0.03 |
ENST00000575129.1 ENST00000398595.3 ENST00000414777.1 |
HMOX2 |
heme oxygenase (decycling) 2 |
| chr5_-_39274617 | 0.03 |
ENST00000510188.1 |
FYB |
FYN binding protein |
| chr1_+_50459990 | 0.03 |
ENST00000448346.1 |
AL645730.2 |
AL645730.2 |
| chr14_+_94385235 | 0.03 |
ENST00000557719.1 ENST00000267594.5 |
FAM181A |
family with sequence similarity 181, member A |
| chr1_+_219347203 | 0.03 |
ENST00000366927.3 |
LYPLAL1 |
lysophospholipase-like 1 |
| chr2_+_33661382 | 0.03 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr4_+_74606223 | 0.03 |
ENST00000307407.3 ENST00000401931.1 |
IL8 |
interleukin 8 |
| chr12_-_11463353 | 0.03 |
ENST00000279575.1 ENST00000535904.1 ENST00000445719.2 |
PRB4 |
proline-rich protein BstNI subfamily 4 |
| chr8_-_20040601 | 0.03 |
ENST00000265808.7 ENST00000522513.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr2_+_145425573 | 0.03 |
ENST00000600064.1 ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr9_+_70856397 | 0.03 |
ENST00000360171.6 |
CBWD3 |
COBW domain containing 3 |
| chr19_+_55385682 | 0.03 |
ENST00000391726.3 |
FCAR |
Fc fragment of IgA, receptor for |
| chr10_-_71169031 | 0.03 |
ENST00000373307.1 |
TACR2 |
tachykinin receptor 2 |
| chr1_+_109255279 | 0.03 |
ENST00000370017.3 |
FNDC7 |
fibronectin type III domain containing 7 |
| chr22_+_19118321 | 0.03 |
ENST00000399635.2 |
TSSK2 |
testis-specific serine kinase 2 |
| chr6_+_46761118 | 0.03 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr4_+_186990298 | 0.03 |
ENST00000296795.3 ENST00000513189.1 |
TLR3 |
toll-like receptor 3 |
| chr8_-_20040638 | 0.03 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr1_+_67632083 | 0.03 |
ENST00000347310.5 ENST00000371002.1 |
IL23R |
interleukin 23 receptor |
| chr2_-_228244013 | 0.03 |
ENST00000304568.3 |
TM4SF20 |
transmembrane 4 L six family member 20 |
| chrX_-_130423386 | 0.03 |
ENST00000370903.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr2_-_179567206 | 0.03 |
ENST00000414766.1 |
TTN |
titin |
| chr1_+_197237352 | 0.03 |
ENST00000538660.1 ENST00000367400.3 ENST00000367399.2 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chrX_-_77150911 | 0.03 |
ENST00000373336.3 |
MAGT1 |
magnesium transporter 1 |
| chr5_+_29143603 | 0.03 |
ENST00000602470.1 ENST00000602801.1 |
CTD-2118P12.1 |
CTD-2118P12.1 |
| chr2_-_231989808 | 0.02 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr13_-_81801115 | 0.02 |
ENST00000567258.1 |
LINC00564 |
long intergenic non-protein coding RNA 564 |
| chr4_-_69817481 | 0.02 |
ENST00000251566.4 |
UGT2A3 |
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr12_-_118628350 | 0.02 |
ENST00000537952.1 ENST00000537822.1 |
TAOK3 |
TAO kinase 3 |
| chr17_-_47723943 | 0.02 |
ENST00000510476.1 ENST00000503676.1 |
SPOP |
speckle-type POZ protein |
| chr17_-_53809473 | 0.02 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr7_+_93535866 | 0.02 |
ENST00000429473.1 ENST00000430875.1 ENST00000428834.1 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr10_-_28623368 | 0.02 |
ENST00000441595.2 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_+_197881592 | 0.02 |
ENST00000367391.1 ENST00000367390.3 |
LHX9 |
LIM homeobox 9 |
| chr4_-_68749699 | 0.02 |
ENST00000545541.1 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr7_-_6866401 | 0.02 |
ENST00000316731.8 |
CCZ1B |
CCZ1 vacuolar protein trafficking and biogenesis associated homolog B (S. cerevisiae) |
| chr15_+_52155001 | 0.02 |
ENST00000544199.1 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chrX_-_109590174 | 0.02 |
ENST00000372054.1 |
GNG5P2 |
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr7_-_5998714 | 0.02 |
ENST00000539903.1 |
RSPH10B |
radial spoke head 10 homolog B (Chlamydomonas) |
| chr13_+_46039037 | 0.02 |
ENST00000349995.5 |
COG3 |
component of oligomeric golgi complex 3 |
| chr6_+_123317116 | 0.02 |
ENST00000275162.5 |
CLVS2 |
clavesin 2 |
| chr3_+_107318157 | 0.02 |
ENST00000406780.1 |
BBX |
bobby sox homolog (Drosophila) |
| chr4_+_88529681 | 0.02 |
ENST00000399271.1 |
DSPP |
dentin sialophosphoprotein |
| chr11_+_89867803 | 0.02 |
ENST00000321955.4 ENST00000525171.1 ENST00000375944.3 |
NAALAD2 |
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr2_+_143635222 | 0.02 |
ENST00000375773.2 ENST00000409512.1 ENST00000410015.2 |
KYNU |
kynureninase |
| chr3_+_195413160 | 0.02 |
ENST00000599448.1 |
LINC00969 |
long intergenic non-protein coding RNA 969 |
| chr1_-_213020991 | 0.02 |
ENST00000332912.3 |
C1orf227 |
chromosome 1 open reading frame 227 |
| chr12_+_9144626 | 0.02 |
ENST00000543895.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr15_+_58430567 | 0.02 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr3_-_139396801 | 0.02 |
ENST00000413939.2 ENST00000339837.5 ENST00000512391.1 |
NMNAT3 |
nicotinamide nucleotide adenylyltransferase 3 |
| chr2_+_234668894 | 0.02 |
ENST00000305208.5 ENST00000608383.1 ENST00000360418.3 |
UGT1A8 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr13_-_33924755 | 0.02 |
ENST00000439831.1 ENST00000567873.1 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr16_+_67927147 | 0.02 |
ENST00000291041.5 |
PSKH1 |
protein serine kinase H1 |
| chr15_+_58430368 | 0.02 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr18_-_22932080 | 0.02 |
ENST00000584787.1 ENST00000361524.3 ENST00000538137.2 |
ZNF521 |
zinc finger protein 521 |
| chr4_-_144940477 | 0.02 |
ENST00000513128.1 ENST00000429670.2 ENST00000502664.1 |
GYPB |
glycophorin B (MNS blood group) |
| chr7_+_93535817 | 0.02 |
ENST00000248572.5 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr4_-_146859787 | 0.02 |
ENST00000508784.1 |
ZNF827 |
zinc finger protein 827 |
| chr7_+_117864708 | 0.02 |
ENST00000357099.4 ENST00000265224.4 ENST00000486422.1 ENST00000417525.1 |
ANKRD7 |
ankyrin repeat domain 7 |
| chr3_-_139396853 | 0.02 |
ENST00000406164.1 ENST00000406824.1 |
NMNAT3 |
nicotinamide nucleotide adenylyltransferase 3 |
| chr3_+_186435065 | 0.02 |
ENST00000287611.2 ENST00000265023.4 |
KNG1 |
kininogen 1 |
| chr2_-_170430277 | 0.02 |
ENST00000438035.1 ENST00000453929.2 |
FASTKD1 |
FAST kinase domains 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0061341 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 0.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.0 | GO:0006788 | heme oxidation(GO:0006788) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.1 | 0.6 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |