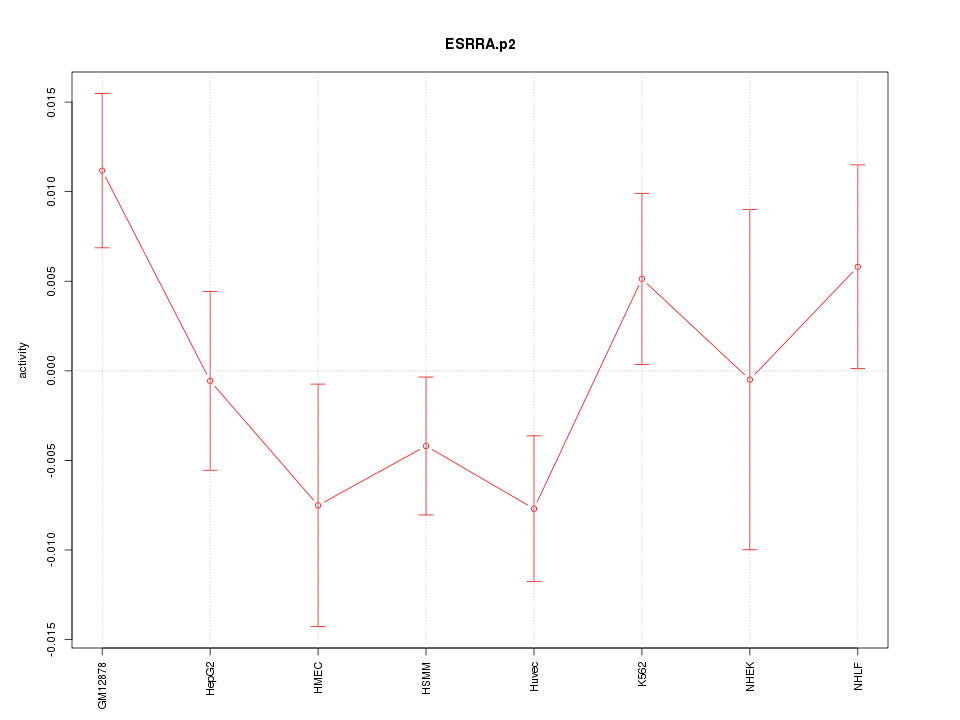
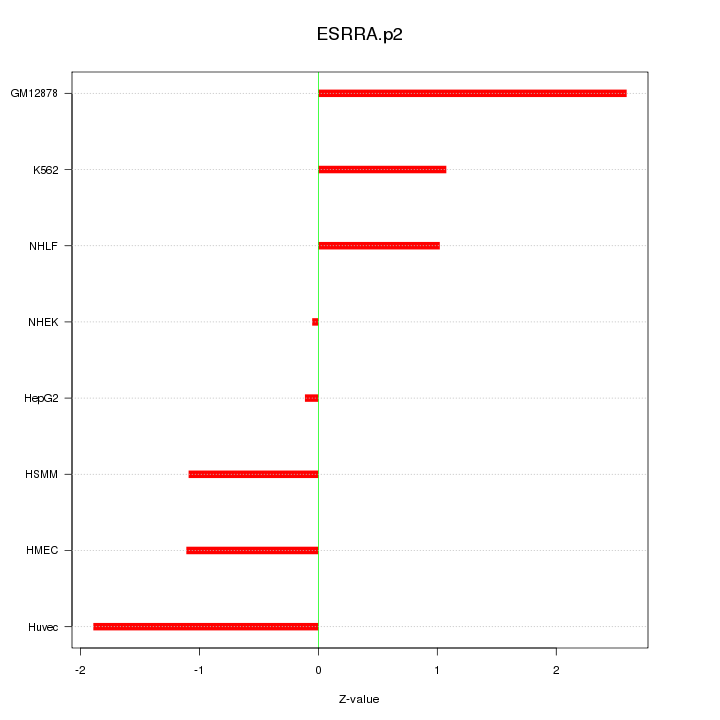
|
chr19_+_540849
|
3.122
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr19_+_1358531
|
1.892
|
NM_018959
NM_170711
|
DAZAP1
|
DAZ associated protein 1
|
|
chr19_+_1358664
|
1.711
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr1_+_29086206
|
1.579
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr19_+_1358604
|
1.475
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr15_+_38520594
|
1.409
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr1_+_29086156
|
1.401
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr5_+_271447
|
1.397
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr12_+_131577229
|
1.389
|
NM_001142641
|
FBRSL1
|
fibrosin-like 1
|
|
chr5_+_271428
|
1.351
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr19_+_1358750
|
1.346
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr19_+_59796858
|
1.345
|
NM_006863
|
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1
|
|
chr5_+_271441
|
1.344
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr6_+_39124534
|
1.254
|
NM_002062
|
GLP1R
|
glucagon-like peptide 1 receptor
|
|
chr1_+_151147646
|
1.230
|
NM_005547
|
IVL
|
involucrin
|
|
chr5_+_271355
|
1.227
|
NM_004168
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr6_+_34314545
|
1.209
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr10_+_44726635
|
1.183
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chr5_+_38481392
|
1.111
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr19_-_4468715
|
1.109
|
NM_001080400
|
PLIN4
|
perilipin 4
|
|
chr1_+_55237193
|
1.097
|
NM_057176
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin)
|
|
chr1_-_19089971
|
1.087
|
NM_001161504
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr6_+_44076281
|
1.077
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr22_+_29151651
|
1.073
|
NM_001003704
NM_016498
|
MTFP1
|
mitochondrial fission process 1
|
|
chr11_-_64284762
|
1.049
|
NM_001164716
NM_005609
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr1_+_29011240
|
1.045
|
NM_000911
|
OPRD1
|
opioid receptor, delta 1
|
|
chr22_-_24088523
|
1.029
|
NM_182492
|
LRP5L
|
low density lipoprotein receptor-related protein 5-like
|
|
chr22_+_22703107
|
1.020
|
NM_001144931
|
LOC391322
|
D-dopachrome tautomerase-like
|
|
chr17_-_70367479
|
1.017
|
NM_000835
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C
|
|
chr10_+_80777248
|
0.987
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr8_+_145221973
|
0.963
|
|
CYC1
|
cytochrome c-1
|
|
chr10_+_80777265
|
0.945
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr16_-_65510230
|
0.943
|
NM_004062
|
CDH16
|
cadherin 16, KSP-cadherin
|
|
chr1_+_15955719
|
0.939
|
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr7_+_45580704
|
0.917
|
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr17_+_7549138
|
0.916
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr1_+_29113589
|
0.912
|
NM_001166006
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr16_-_73842884
|
0.906
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr16_+_55456619
|
0.903
|
NM_000339
NM_001126107
NM_001126108
|
SLC12A3
|
solute carrier family 12 (sodium/chloride transporters), member 3
|
|
chr14_-_104507862
|
0.902
|
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr7_+_45580606
|
0.897
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr11_-_780103
|
0.891
|
NM_016564
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr19_+_45807909
|
0.842
|
|
|
|
|
chrX_+_69371229
|
0.832
|
NM_001013579
|
AWAT1
|
acyl-CoA wax alcohol acyltransferase 1
|
|
chr22_+_38903825
|
0.831
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr9_-_116190038
|
0.827
|
|
AKNA
|
AT-hook transcription factor
|
|
chr11_-_780065
|
0.818
|
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr3_+_130730171
|
0.813
|
NM_000539
|
RHO
|
rhodopsin
|
|
chr15_+_41597097
|
0.801
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr7_+_97679501
|
0.798
|
NM_177455
|
BHLHA15
|
basic helix-loop-helix family, member a15
|
|
chr7_-_158304062
|
0.792
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr15_+_62991106
|
0.780
|
NM_182703
|
ANKDD1A
|
ankyrin repeat and death domain containing 1A
|
|
chr5_-_271235
|
0.760
|
NM_145265
|
CCDC127
|
coiled-coil domain containing 127
|
|
chr2_-_128792515
|
0.750
|
NM_004807
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1
|
|
chr1_-_150065176
|
0.727
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr7_+_116238309
|
0.723
|
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2
|
|
chr19_+_1199548
|
0.716
|
NM_177401
|
MIDN
|
midnolin
|
|
chr11_+_65815940
|
0.706
|
NM_153266
|
TMEM151A
|
transmembrane protein 151A
|
|
chr2_+_119905812
|
0.698
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr6_+_37081387
|
0.698
|
NM_173558
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2
|
|
chr11_+_65140207
|
0.693
|
NM_032223
|
PCNXL3
|
pecanex-like 3 (Drosophila)
|
|
chr12_+_6846955
|
0.686
|
|
TPI1
|
triosephosphate isomerase 1
|
|
chr12_+_6846844
|
0.686
|
NM_001159287
NM_000365
|
TPI1
|
triosephosphate isomerase 1
|
|
chr8_-_139995417
|
0.683
|
NM_152888
|
COL22A1
|
collagen, type XXII, alpha 1
|
|
chr19_+_3136860
|
0.676
|
NM_020170
|
NCLN
|
nicalin
|
|
chr1_+_151655623
|
0.671
|
NM_176823
|
S100A7A
|
S100 calcium binding protein A7A
|
|
chr1_+_226337425
|
0.648
|
NM_001024227
NM_001024228
|
ARF1
|
ADP-ribosylation factor 1
|
|
chr14_-_105245959
|
0.648
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr19_-_60360768
|
0.645
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr11_-_18704294
|
0.643
|
NM_173588
|
IGSF22
|
immunoglobulin superfamily, member 22
|
|
chr22_+_48740432
|
0.634
|
|
PIM3
|
pim-3 oncogene
|
|
chr3_-_42719322
|
0.630
|
NM_020707
|
HHATL
|
hedgehog acyltransferase-like
|
|
chr9_+_37640996
|
0.629
|
NM_014907
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chrX_+_48505097
|
0.616
|
NM_001080489
|
GLOD5
|
glyoxalase domain containing 5
|
|
chr8_+_145221924
|
0.616
|
NM_001916
|
CYC1
|
cytochrome c-1
|
|
chr9_-_130006309
|
0.616
|
NM_001131015
NM_012127
|
CIZ1
|
CDKN1A interacting zinc finger protein 1
|
|
chr22_+_18485145
|
0.612
|
|
RANBP1
|
RAN binding protein 1
|
|
chr20_+_39402834
|
0.612
|
NM_022896
|
LPIN3
|
lipin 3
|
|
chr14_+_99220349
|
0.609
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr8_+_145221966
|
0.599
|
|
CYC1
|
cytochrome c-1
|
|
chr8_-_145099963
|
0.595
|
NM_201379
|
PLEC
|
plectin
|
|
chr16_-_65984836
|
0.595
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr12_+_103677388
|
0.593
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr12_+_121233615
|
0.593
|
NM_001098519
|
LRRC43
|
leucine rich repeat containing 43
|
|
chr11_+_65815911
|
0.587
|
|
TMEM151A
|
transmembrane protein 151A
|
|
chr14_+_104523660
|
0.583
|
NM_174891
|
C14orf79
|
chromosome 14 open reading frame 79
|
|
chr15_+_73281288
|
0.582
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr1_+_38032045
|
0.580
|
NM_001031740
NM_001113482
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr17_-_34014390
|
0.577
|
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr20_-_61600834
|
0.564
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chrX_-_13744933
|
0.564
|
|
GPM6B
|
glycoprotein M6B
|
|
chr7_+_138859213
|
0.560
|
NM_001080511
|
CLEC2L
|
C-type lectin domain family 2, member L
|
|
chr9_-_129719031
|
0.553
|
NM_175039
NM_175040
|
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr17_-_70430839
|
0.549
|
NM_173477
|
USH1G
|
Usher syndrome 1G (autosomal recessive)
|
|
chr19_-_54250808
|
0.548
|
NM_033142
|
CGB7
|
chorionic gonadotropin, beta polypeptide 7
|
|
chr19_+_2200112
|
0.545
|
NM_000479
|
AMH
|
anti-Mullerian hormone
|
|
chr17_-_31332568
|
0.545
|
NM_004590
|
CCL16
|
chemokine (C-C motif) ligand 16
|
|
chr19_+_7734034
|
0.545
|
NM_001144904
NM_001144905
NM_001144906
NM_001144907
NM_001144908
NM_001144909
NM_001144910
NM_001144911
NM_014257
|
CLEC4M
|
C-type lectin domain family 4, member M
|
|
chr22_+_18485095
|
0.541
|
|
RANBP1
|
RAN binding protein 1
|
|
chr3_+_180805266
|
0.538
|
NM_002492
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa
|
|
chr22_+_18485076
|
0.536
|
|
RANBP1
|
RAN binding protein 1
|
|
chr1_-_1679940
|
0.535
|
NM_001198995
|
NADK
|
NAD kinase
|
|
chr10_+_80777234
|
0.535
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr15_+_73281286
|
0.533
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr16_+_658081
|
0.530
|
NM_138769
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr2_-_128149112
|
0.530
|
NM_001136037
NM_001161403
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr9_+_130005429
|
0.528
|
NM_001005336
NM_004408
|
DNM1
|
dynamin 1
|
|
chr9_+_130005489
|
0.527
|
|
DNM1
|
dynamin 1
|
|
chr10_+_75427853
|
0.527
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr10_+_80777199
|
0.526
|
NM_005729
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr9_+_130005487
|
0.525
|
|
DNM1
|
dynamin 1
|
|
chr17_-_8756524
|
0.524
|
NM_014308
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5
|
|
chr6_-_2785682
|
0.520
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr22_+_18484810
|
0.512
|
NM_002882
|
RANBP1
|
RAN binding protein 1
|
|
chr15_+_73281249
|
0.510
|
NM_015492
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr3_-_180805104
|
0.510
|
NM_020409
NM_177988
|
MRPL47
|
mitochondrial ribosomal protein L47
|
|
chr1_+_32530294
|
0.507
|
NM_004964
|
HDAC1
|
histone deacetylase 1
|
|
chr1_+_243384868
|
0.503
|
NM_018012
|
KIF26B
|
kinesin family member 26B
|
|
chr7_+_97748959
|
0.502
|
|
BRI3
|
brain protein I3
|
|
chr4_+_7096123
|
0.502
|
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr1_+_32530298
|
0.500
|
|
HDAC1
|
histone deacetylase 1
|
|
chr1_+_32530289
|
0.495
|
|
HDAC1
|
histone deacetylase 1
|
|
chr1_+_9338531
|
0.484
|
|
|
|
|
chr19_+_44308224
|
0.484
|
NM_001014831
NM_001014832
NM_001014834
NM_001014835
NM_005884
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr19_+_17923110
|
0.480
|
NM_002248
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1
|
|
chr12_+_119360274
|
0.476
|
NM_004373
|
COX6A1
|
cytochrome c oxidase subunit VIa polypeptide 1
|
|
chr14_-_102045849
|
0.474
|
NM_152326
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr4_+_7245218
|
0.473
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr3_+_9720509
|
0.472
|
NM_153635
|
CPNE9
|
copine family member IX
|
|
chr10_-_97190885
|
0.471
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr16_+_66464492
|
0.469
|
NM_014329
|
EDC4
|
enhancer of mRNA decapping 4
|
|
chr15_-_38417564
|
0.468
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr19_+_4134350
|
0.461
|
NM_133475
|
ANKRD24
|
ankyrin repeat domain 24
|
|
chr19_+_59866217
|
0.450
|
|
LILRB4
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4
|
|
chr3_-_51971915
|
0.447
|
NM_033010
|
PCBP4
|
poly(rC) binding protein 4
|
|
chrX_-_53002900
|
0.447
|
NM_014138
|
FAM156A
|
family with sequence similarity 156, member A
|
|
chr13_+_112349358
|
0.446
|
NM_207440
|
C13orf35
|
chromosome 13 open reading frame 35
|
|
chr22_+_28032950
|
0.445
|
NM_006478
NM_152236
NM_152237
|
GAS2L1
|
growth arrest-specific 2 like 1
|
|
chr22_+_18485068
|
0.435
|
|
RANBP1
|
RAN binding protein 1
|
|
chr14_+_69021149
|
0.433
|
NM_001161498
|
UPF0639
|
UPF0639 protein
|
|
chr7_-_100630912
|
0.427
|
NM_178176
|
MOGAT3
|
monoacylglycerol O-acyltransferase 3
|
|
chr13_+_97856148
|
0.426
|
|
|
|
|
chr16_+_4370879
|
0.422
|
|
VASN
|
vasorin
|
|
chr7_+_150413814
|
0.418
|
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr1_-_13263334
|
0.418
|
NM_001012276
|
PRAMEF8
PRAMEF7
|
PRAME family member 8
PRAME family member 7
|
|
chr19_+_54238913
|
0.417
|
NM_033043
|
CGB
CGB5
|
chorionic gonadotropin, beta polypeptide
chorionic gonadotropin, beta polypeptide 5
|
|
chr19_-_1464187
|
0.413
|
NM_213604
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr16_+_658158
|
0.413
|
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr14_-_102045749
|
0.409
|
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr11_+_67554653
|
0.409
|
NM_002496
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase)
|
|
chr1_-_33110668
|
0.405
|
NM_001171941
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr4_-_40212670
|
0.405
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr19_+_59297953
|
0.403
|
NM_004542
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
|
|
chr16_+_30844707
|
0.402
|
|
FBXL19
|
F-box and leucine-rich repeat protein 19
|
|
chr2_-_172675873
|
0.401
|
|
DLX2
|
distal-less homeobox 2
|
|
chr2_-_27339278
|
0.401
|
NM_003459
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr17_+_45008473
|
0.401
|
|
NXPH3
|
neurexophilin 3
|
|
chr17_+_45778381
|
0.398
|
NM_022167
|
XYLT2
|
xylosyltransferase II
|
|
chr19_+_38867273
|
0.398
|
NM_022467
|
CHST8
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8
|
|
chr19_-_12028126
|
0.394
|
NM_001080404
|
ZNF878
|
zinc finger protein 878
|
|
chr11_+_43920385
|
0.394
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr16_-_2663375
|
0.391
|
|
|
|
|
chr19_+_2279679
|
0.390
|
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr22_+_35586950
|
0.388
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr1_+_20269239
|
0.388
|
NM_000929
|
PLA2G5
|
phospholipase A2, group V
|
|
chr15_-_76210611
|
0.386
|
NM_006383
|
CIB2
|
calcium and integrin binding family member 2
|
|
chr5_+_176807931
|
0.386
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr20_-_45848684
|
0.385
|
NM_001161841
NM_198596
|
SULF2
|
sulfatase 2
|
|
chr16_+_2528022
|
0.384
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr4_-_1827475
|
0.382
|
NM_012318
|
LETM1
|
leucine zipper-EF-hand containing transmembrane protein 1
|
|
chr11_+_1817294
|
0.381
|
NM_001145829
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr20_-_1922393
|
0.380
|
NM_001190900
|
PDYN
|
prodynorphin
|
|
chr20_-_3696344
|
0.378
|
NM_001039140
|
C20orf27
|
chromosome 20 open reading frame 27
|
|
chr19_+_2279685
|
0.378
|
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chrX_-_44087778
|
0.378
|
NM_025184
|
EFHC2
|
EF-hand domain (C-terminal) containing 2
|
|
chr1_+_17719405
|
0.377
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr11_-_75769514
|
0.374
|
NM_004705
|
PRKRIR
|
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor)
|
|
chr1_+_234625338
|
0.372
|
NM_080738
|
EDARADD
|
EDAR-associated death domain
|
|
chr16_+_4614820
|
0.372
|
NM_001142289
NM_001142290
NM_001142291
NM_015246
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr19_-_1463997
|
0.371
|
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr16_+_2528001
|
0.370
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr6_+_36773605
|
0.370
|
|
RAB44
|
RAB44, member RAS oncogene family
|
|
chr1_-_13484119
|
0.366
|
NM_001012276
|
PRAMEF8
PRAMEF7
|
PRAME family member 8
PRAME family member 7
|
|
chr17_+_77289775
|
0.365
|
NM_012140
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10
|
|
chr9_-_137992967
|
0.364
|
NM_016172
|
UBAC1
|
UBA domain containing 1
|
|
chr22_+_18485057
|
0.363
|
|
RANBP1
|
RAN binding protein 1
|
|
chr8_-_131098015
|
0.360
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr16_+_2527952
|
0.359
|
NM_002613
NM_031268
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chrX_+_152682772
|
0.359
|
NM_001163257
NM_005393
|
PLXNB3
|
plexin B3
|
|
chr17_+_35075124
|
0.357
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chr16_+_4614959
|
0.352
|
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr19_+_62434188
|
0.352
|
NM_001015879
NM_003160
NM_001015878
|
AURKC
|
aurora kinase C
|
|
chr8_-_145090892
|
0.351
|
NM_201381
|
PLEC
|
plectin
|
|
chr17_-_45619038
|
0.347
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr3_-_10003512
|
0.344
|
NM_018447
|
TMEM111
|
transmembrane protein 111
|
|
chr22_-_43040063
|
0.343
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr1_-_202726068
|
0.343
|
NM_002646
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr11_+_111462821
|
0.342
|
|
SDHD
|
succinate dehydrogenase complex, subunit D, integral membrane protein
|
|
chr11_+_66381077
|
0.342
|
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr13_+_113587674
|
0.341
|
|
GAS6
|
growth arrest-specific 6
|