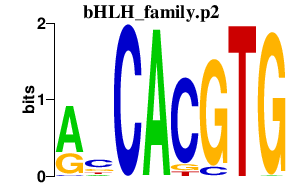
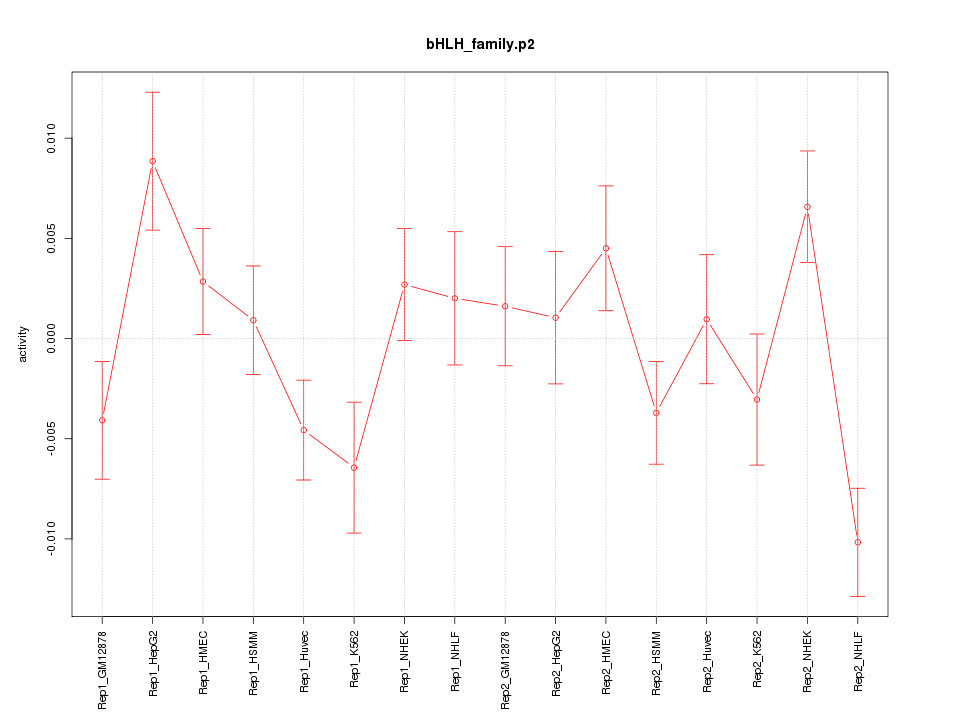
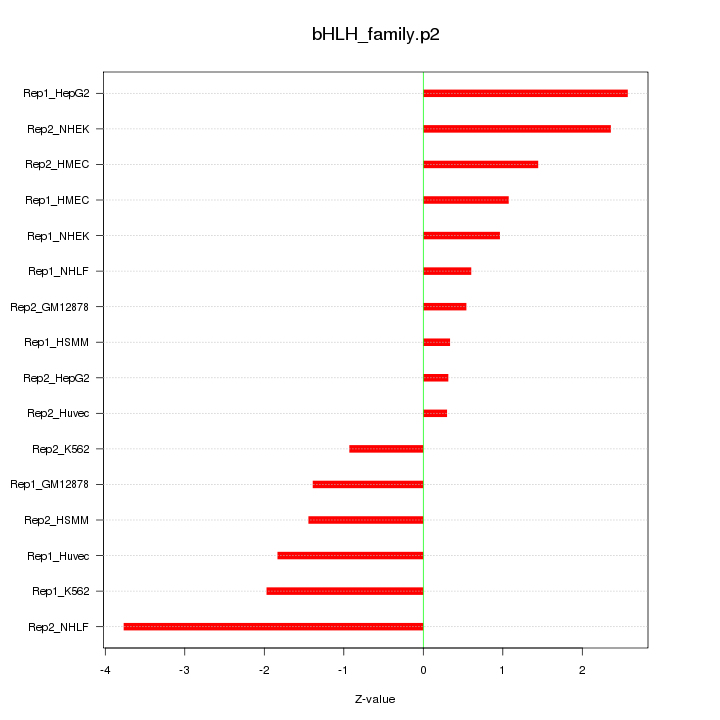
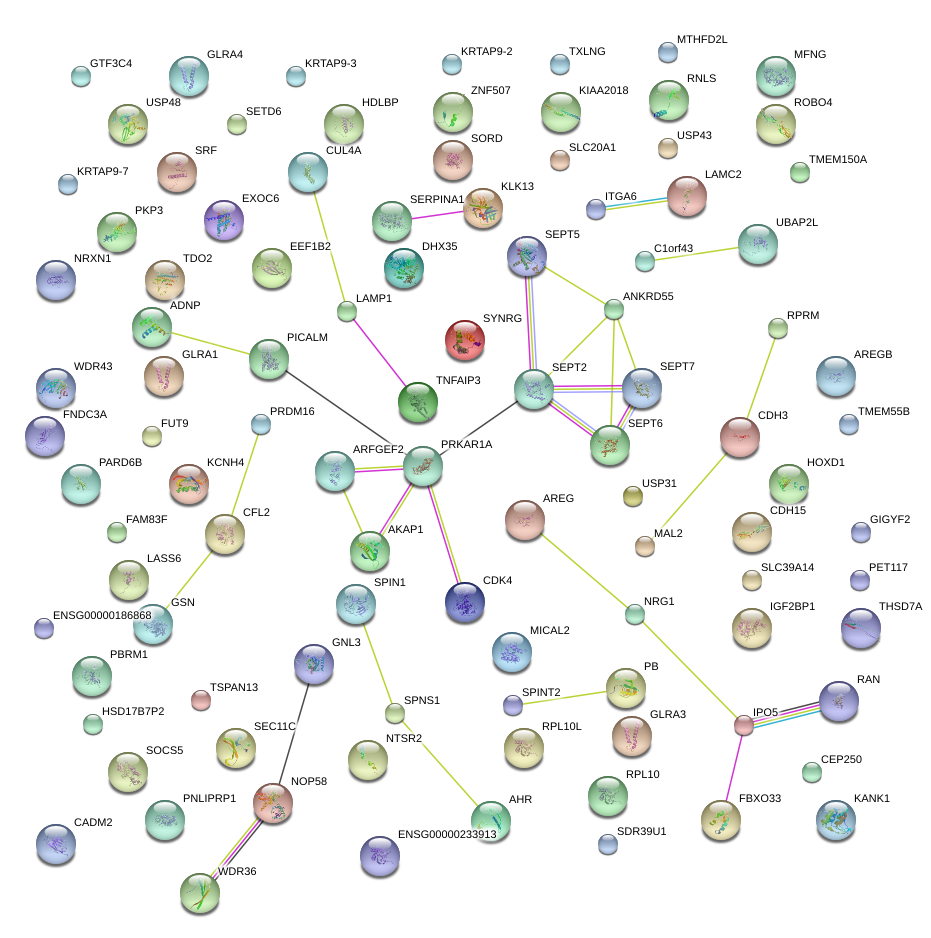
|
chr22_+_38720881
|
3.054
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr8_+_120289735
|
2.610
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr6_+_138230045
|
2.232
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr20_+_46971618
|
1.794
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr8_+_32525269
|
1.509
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr2_+_28971007
|
1.308
|
NM_015131
|
WDR43
|
WD repeat domain 43
|
|
chr6_+_96570565
|
1.281
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr17_+_44429748
|
1.180
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr2_+_46779553
|
1.165
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr8_+_22281118
|
1.133
|
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr2_+_202838725
|
1.124
|
NM_015934
|
NOP58
|
NOP58 ribonucleoprotein homolog (yeast)
|
|
chr2_+_113119985
|
1.106
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr2_+_46779827
|
1.065
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr14_-_46190690
|
1.020
|
NM_080746
|
RPL10L
|
ribosomal protein L10-like
|
|
chr11_-_85457736
|
1.020
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr16_+_67236699
|
1.004
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr8_+_22280942
|
1.004
|
NM_001135153
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr17_-_33043514
|
1.000
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr5_+_110455768
|
0.988
|
NM_139281
|
WDR36
|
WD repeat domain 36
|
|
chr2_+_206732859
|
0.971
|
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr14_-_34253473
|
0.971
|
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr14_-_93919305
|
0.970
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr20_+_37024383
|
0.967
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr17_+_9489578
|
0.942
|
NM_153210
|
USP43
|
ubiquitin specific peptidase 43
|
|
chr17_+_52518098
|
0.938
|
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr2_+_202838680
|
0.933
|
|
NOP58
|
NOP58 ribonucleoprotein homolog (yeast)
|
|
chr11_-_85457753
|
0.888
|
NM_001008660
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr1_-_152459506
|
0.881
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr13_+_112999462
|
0.866
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr2_-_50428395
|
0.864
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr7_+_17304707
|
0.862
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr8_+_22280731
|
0.862
|
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr5_+_110455961
|
0.853
|
|
WDR36
|
WD repeat domain 36
|
|
chr2_+_169021001
|
0.852
|
|
LASS6
|
LAG1 homolog, ceramide synthase 6
|
|
chr1_-_152459667
|
0.850
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr19_+_37528340
|
0.843
|
NM_001136156
NM_014910
|
ZNF507
|
zinc finger protein 507
|
|
chr11_+_384199
|
0.834
|
NM_007183
|
PKP3
|
plakophilin 3
|
|
chr8_+_22280703
|
0.832
|
NM_001128431
NM_001135154
NM_015359
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr2_+_169021039
|
0.827
|
NM_203463
|
LASS6
|
LAG1 homolog, ceramide synthase 6
|
|
chr16_-_23068010
|
0.825
|
NM_020718
|
USP31
|
ubiquitin specific peptidase 31
|
|
chr4_-_175986952
|
0.816
|
NM_001042543
NM_006529
|
GLRA3
|
glycine receptor, alpha 3
|
|
chr3_-_52694554
|
0.810
|
NM_018313
|
PBRM1
|
polybromo 1
|
|
chr17_-_37586716
|
0.809
|
NM_012285
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4
|
|
chr11_+_384238
|
0.796
|
|
PKP3
|
plakophilin 3
|
|
chr7_-_11838260
|
0.793
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr13_+_97426713
|
0.791
|
|
IPO5
|
importin 5
|
|
chr2_+_206732575
|
0.790
|
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr6_-_138231006
|
0.771
|
|
|
|
|
chrX_-_114703273
|
0.769
|
|
|
|
|
chr1_+_152459918
|
0.765
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr20_-_48980858
|
0.761
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr4_+_75529716
|
0.759
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr2_-_11727705
|
0.747
|
NM_012344
|
NTSR2
|
neurotensin receptor 2
|
|
chr3_+_85090721
|
0.739
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chrX_+_16714452
|
0.737
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr13_+_112999538
|
0.733
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr20_+_48781457
|
0.720
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr16_+_57106883
|
0.703
|
NM_001160305
NM_024860
|
SETD6
|
SET domain containing 6
|
|
chr2_+_241903395
|
0.702
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr18_+_54958095
|
0.689
|
NM_033280
|
SEC11C
|
SEC11 homolog C (S. cerevisiae)
|
|
chr13_+_112911927
|
0.686
|
NM_001008895
|
CUL4A
|
cullin 4A
|
|
chr11_-_85457469
|
0.683
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr20_-_604443
|
0.681
|
|
|
|
|
chr2_-_154043403
|
0.678
|
NM_019845
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chr10_+_118340460
|
0.676
|
NM_006229
|
PNLIPRP1
|
pancreatic lipase-related protein 1
|
|
chr4_+_75242671
|
0.668
|
NM_001144978
|
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like
|
|
chr2_+_233270286
|
0.667
|
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr2_-_241903657
|
0.667
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr13_+_48582389
|
0.663
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr17_+_64020120
|
0.661
|
NM_002734
NM_212472
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr12_+_129922570
|
0.659
|
|
RAN
|
RAN, member RAS oncogene family
|
|
chr9_+_134535210
|
0.658
|
|
GTF3C4
|
general transcription factor IIIC, polypeptide 4, 90kDa
|
|
chr13_+_112911826
|
0.657
|
|
CUL4A
|
cullin 4A
|
|
chr14_-_19999475
|
0.656
|
NM_001100814
NM_144568
|
TMEM55B
|
transmembrane protein 55B
|
|
chr7_+_16759845
|
0.651
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr2_-_85683268
|
0.647
|
NM_001031738
|
TMEM150A
|
transmembrane protein 150A
|
|
chr2_-_74915722
|
0.642
|
|
|
|
|
chr12_-_56432385
|
0.640
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr14_-_23981493
|
0.638
|
NM_020195
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1
|
|
chr3_+_52694975
|
0.637
|
NM_014366
NM_206825
NM_206826
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr17_+_36636404
|
0.637
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr14_-_38971370
|
0.636
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr17_+_70256518
|
0.635
|
|
|
|
|
chr10_+_94598204
|
0.632
|
NM_019053
|
EXOC6
|
exocyst complex component 6
|
|
chr19_+_43447262
|
0.627
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr20_+_18066473
|
0.626
|
NM_001164811
|
PET117
|
cytochrome c oxidase assembly factor-like
|
|
chr9_+_696744
|
0.625
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr15_+_43102593
|
0.625
|
NM_003104
|
SORD
|
sorbitol dehydrogenase
|
|
chr16_+_67236730
|
0.624
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr2_+_233270275
|
0.624
|
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr1_+_181422016
|
0.624
|
|
LAMC2
|
laminin, gamma 2
|
|
chr2_+_176761552
|
0.619
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr17_+_64020155
|
0.617
|
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr16_+_28893525
|
0.614
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr11_+_12088639
|
0.608
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr2_-_241903787
|
0.607
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr10_-_90332927
|
0.605
|
NM_001031709
NM_018363
|
RNLS
|
renalase, FAD-dependent amine oxidase
|
|
chr22_-_36212160
|
0.605
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr2_+_173000590
|
0.604
|
|
ITGA6
|
integrin, alpha 6
|
|
chr20_+_33506322
|
0.603
|
NM_007186
|
CEP250
|
centrosomal protein 250kDa
|
|
chr12_-_56432375
|
0.602
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr17_+_46692908
|
0.599
|
|
UTP18
|
UTP18, small subunit (SSU) processome component, homolog (yeast)
|
|
chr14_+_44622916
|
0.596
|
NM_017922
|
PRPF39
|
PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae)
|
|
chr14_+_23855291
|
0.587
|
|
LTB4R
|
leukotriene B4 receptor
|
|
chr1_-_152459697
|
0.587
|
NM_001098616
NM_015449
NM_138740
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr12_-_56432377
|
0.586
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr19_+_6690697
|
0.585
|
NM_004240
|
TRIP10
|
thyroid hormone receptor interactor 10
|
|
chr2_+_233270241
|
0.585
|
NM_001103146
NM_001103147
NM_001103148
NM_015575
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr18_+_75967902
|
0.583
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr9_-_134535148
|
0.582
|
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31
|
|
chr4_+_75699652
|
0.582
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr20_-_604822
|
0.581
|
NM_033129
|
SCRT2
|
scratch homolog 2, zinc finger protein (Drosophila)
|
|
chr7_+_43118721
|
0.576
|
NM_015052
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr10_-_104463947
|
0.575
|
NM_004311
|
ARL3
|
ADP-ribosylation factor-like 3
|
|
chr17_+_64020159
|
0.575
|
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr5_+_41940216
|
0.575
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr12_+_129922589
|
0.572
|
|
RAN
|
RAN, member RAS oncogene family
|
|
chrX_+_30581523
|
0.572
|
|
GK
|
glycerol kinase
|
|
chr17_+_64019693
|
0.563
|
NM_212471
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr13_+_112999590
|
0.561
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr12_-_131816604
|
0.559
|
|
ANKLE2
|
ankyrin repeat and LEM domain containing 2
|
|
chr20_+_39091026
|
0.559
|
|
TOP1
|
topoisomerase (DNA) I
|
|
chr14_+_23700261
|
0.555
|
NM_006084
|
IRF9
|
interferon regulatory factor 9
|
|
chr16_+_273118
|
0.554
|
NM_006849
|
PDIA2
|
protein disulfide isomerase family A, member 2
|
|
chr1_+_237616457
|
0.554
|
|
CHRM3
|
cholinergic receptor, muscarinic 3
|
|
chr19_+_43446937
|
0.553
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr2_-_131567458
|
0.551
|
NM_001009993
|
FAM168B
|
family with sequence similarity 168, member B
|
|
chr20_-_41788975
|
0.550
|
NM_001008901
NM_176791
|
GTSF1L
|
gametocyte specific factor 1-like
|
|
chr18_-_45241014
|
0.549
|
NM_017653
|
DYM
|
dymeclin
|
|
chr12_+_32546243
|
0.548
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr19_+_43447078
|
0.547
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr16_+_55523526
|
0.546
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr17_+_64019748
|
0.543
|
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr7_+_94130673
|
0.540
|
|
PEG10
|
paternally expressed 10
|
|
chr13_-_49597776
|
0.535
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr10_-_32257693
|
0.530
|
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr14_-_23727826
|
0.530
|
|
IPO4
|
importin 4
|
|
chr2_+_227898163
|
0.530
|
|
MFF
|
mitochondrial fission factor
|
|
chr20_+_39090875
|
0.527
|
NM_003286
|
TOP1
|
topoisomerase (DNA) I
|
|
chr2_+_190014403
|
0.526
|
NM_032168
|
WDR75
|
WD repeat domain 75
|
|
chr12_-_119826073
|
0.525
|
|
SPPL3
|
signal peptide peptidase 3
|
|
chr17_+_32380287
|
0.525
|
NM_012138
|
AATF
|
apoptosis antagonizing transcription factor
|
|
chr17_-_54538816
|
0.521
|
|
TRIM37
|
tripartite motif containing 37
|
|
chr2_+_218992053
|
0.521
|
NM_007127
|
VIL1
|
villin 1
|
|
chr9_+_6747919
|
0.519
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr2_-_217267742
|
0.519
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr17_+_70256397
|
0.516
|
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr7_+_94374862
|
0.515
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr17_-_54538898
|
0.515
|
|
TRIM37
|
tripartite motif containing 37
|
|
chr6_+_143423738
|
0.513
|
|
AIG1
|
androgen-induced 1
|
|
chr5_+_80292213
|
0.512
|
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr18_+_75968133
|
0.512
|
|
ADNP2
|
ADNP homeobox 2
|
|
chr6_+_143423723
|
0.511
|
|
AIG1
|
androgen-induced 1
|
|
chr17_-_54538956
|
0.510
|
NM_001005207
NM_015294
|
TRIM37
|
tripartite motif containing 37
|
|
chr7_-_86526880
|
0.510
|
NM_001142749
|
KIAA1324L
|
KIAA1324-like
|
|
chr10_+_103882776
|
0.510
|
NM_015062
|
PPRC1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1
|
|
chr1_+_42920946
|
0.508
|
|
YBX1
|
Y box binding protein 1
|
|
chr6_+_7052985
|
0.508
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr17_+_70256341
|
0.504
|
NM_004252
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr1_+_181421984
|
0.504
|
|
LAMC2
|
laminin, gamma 2
|
|
chr10_+_69314347
|
0.503
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr9_+_71009746
|
0.501
|
NM_001170415
NM_001170416
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr17_+_70256389
|
0.499
|
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr20_+_5934761
|
0.498
|
|
CRLS1
|
cardiolipin synthase 1
|
|
chr17_+_70256370
|
0.494
|
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr21_+_33844793
|
0.491
|
|
SON
|
SON DNA binding protein
|
|
chr13_+_77170470
|
0.490
|
NM_001040153
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr9_-_124707314
|
0.489
|
NM_001100588
NM_018835
|
RC3H2
|
ring finger and CCCH-type domains 2
|
|
chrX_-_107866262
|
0.488
|
NM_003604
|
IRS4
|
insulin receptor substrate 4
|
|
chr12_+_6703686
|
0.487
|
NM_001164093
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr20_+_32311831
|
0.484
|
NM_001672
|
ASIP
|
agouti signaling protein
|
|
chr12_+_70434920
|
0.482
|
NM_014999
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr11_+_32993973
|
0.482
|
NM_001077242
|
DEPDC7
|
DEP domain containing 7
|
|
chr2_+_42249896
|
0.480
|
NM_001145076
NM_019063
|
EML4
|
echinoderm microtubule associated protein like 4
|
|
chr3_-_172109119
|
0.476
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr20_+_5934560
|
0.476
|
NM_019095
|
CRLS1
|
cardiolipin synthase 1
|
|
chr7_+_94130752
|
0.475
|
|
PEG10
|
paternally expressed 10
|
|
chr14_-_38971049
|
0.471
|
|
FBXO33
|
F-box protein 33
|
|
chr20_+_5934964
|
0.470
|
|
CRLS1
|
cardiolipin synthase 1
|
|
chr22_+_27609889
|
0.470
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr10_-_124758178
|
0.467
|
NM_022466
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus)
|
|
chr20_+_13924263
|
0.460
|
|
MACROD2
|
MACRO domain containing 2
|
|
chr16_+_85983447
|
0.459
|
|
MAP1LC3B
|
microtubule-associated protein 1 light chain 3 beta
|
|
chr14_+_38806280
|
0.457
|
|
CTAGE5
|
CTAGE family, member 5
|
|
chr17_-_71362971
|
0.456
|
NM_012478
|
WBP2
|
WW domain binding protein 2
|
|
chr5_-_65053630
|
0.455
|
NM_019072
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta
|
|
chr13_+_32488477
|
0.455
|
NM_004795
|
KL
|
klotho
|
|
chrX_+_55760774
|
0.455
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr2_+_215885063
|
0.454
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr6_+_7052798
|
0.453
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr22_-_37425937
|
0.450
|
NM_014876
|
JOSD1
|
Josephin domain containing 1
|
|
chr22_+_40027529
|
0.450
|
|
ZC3H7B
|
zinc finger CCCH-type containing 7B
|
|
chr1_+_149779384
|
0.449
|
NM_001126337
NM_020127
|
TUFT1
|
tuftelin 1
|
|
chr13_-_27572646
|
0.449
|
NM_004119
|
FLT3
|
fms-related tyrosine kinase 3
|
|
chr10_+_75580914
|
0.445
|
NM_006721
|
ADK
|
adenosine kinase
|
|
chr2_+_208284484
|
0.442
|
NM_001142300
NM_152523
|
CCNYL1
|
cyclin Y-like 1
|
|
chr12_+_19173669
|
0.441
|
NM_001143821
NM_001190860
NM_019012
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5
|
|
chr9_+_70926205
|
0.440
|
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr2_+_55313434
|
0.438
|
|
RPS27A
|
ribosomal protein S27a
|
|
chr2_+_215885053
|
0.437
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|