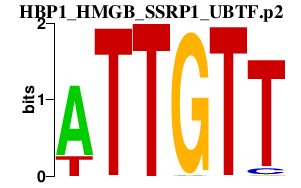
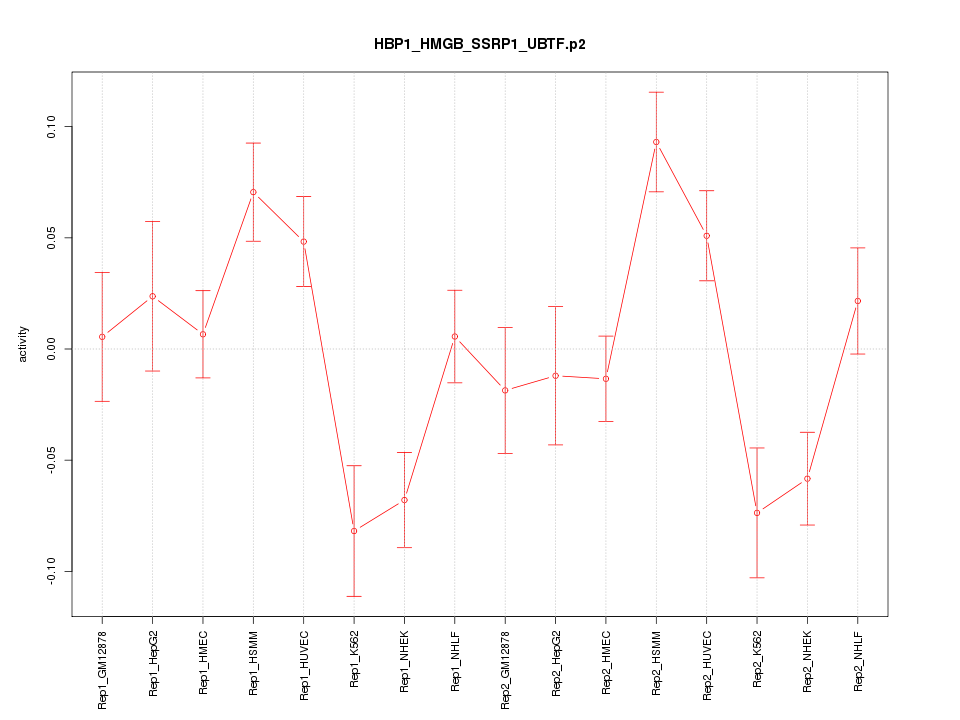
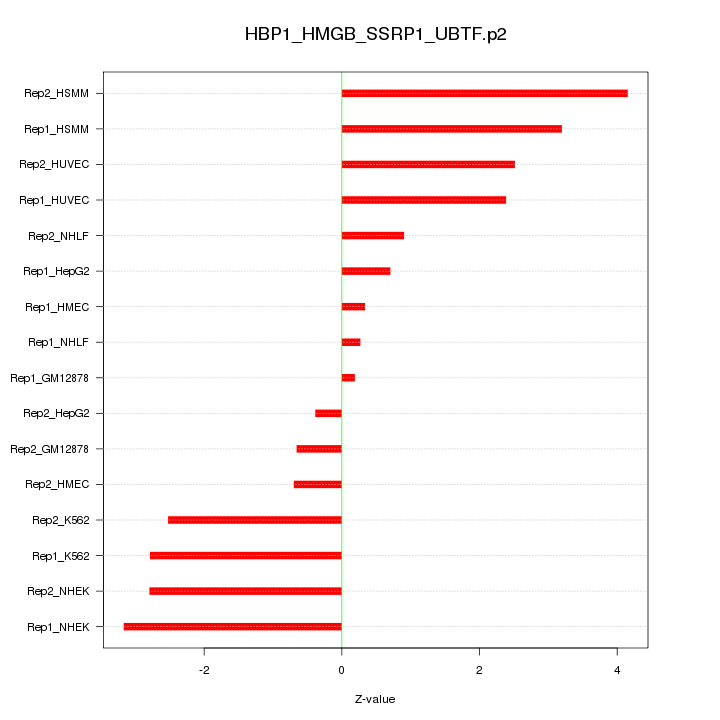
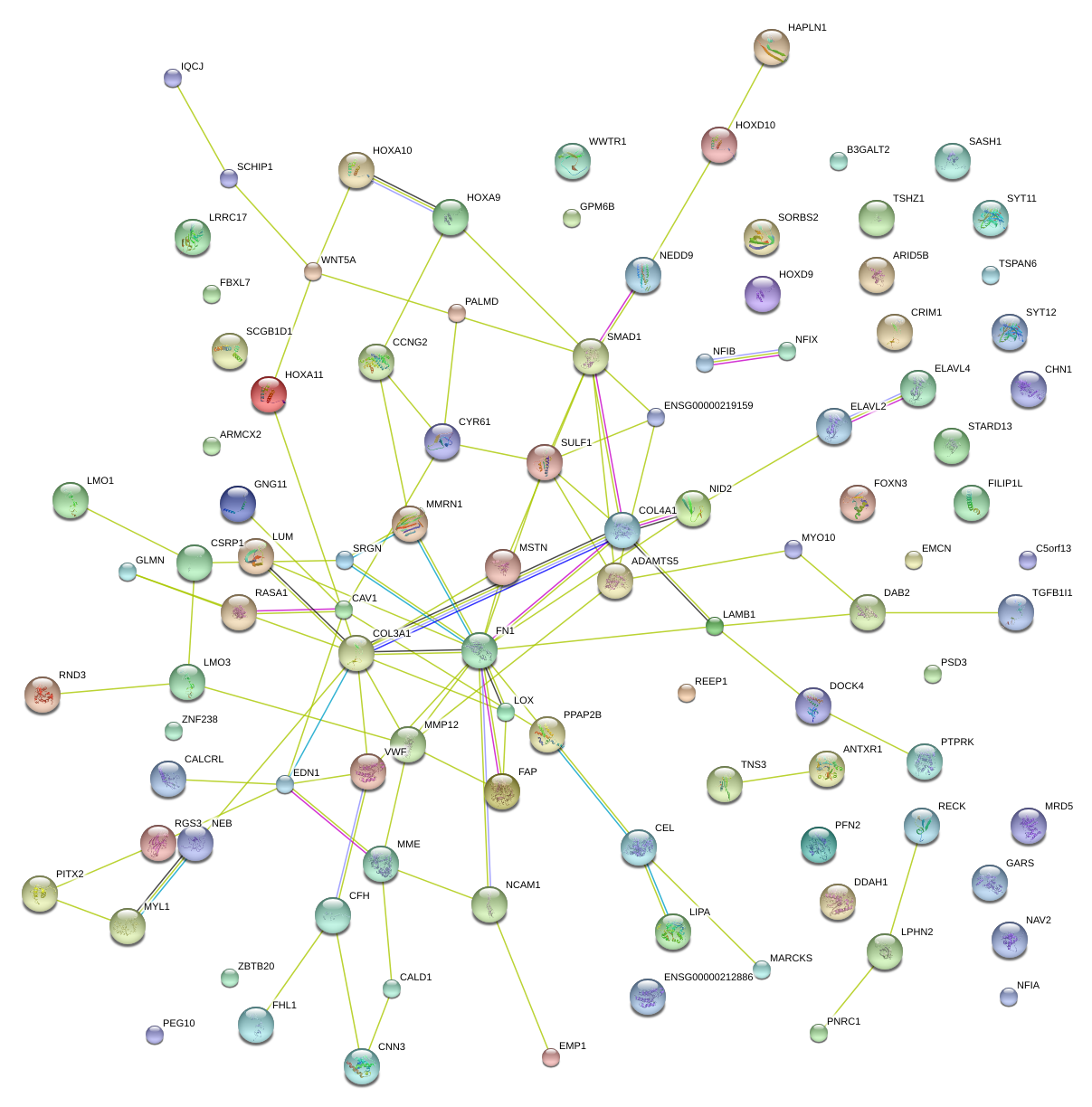
|
chr5_-_111119846
|
8.684
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_+_134114957
|
6.714
|
|
CALD1
|
caldesmon 1
|
|
chr7_+_102340579
|
6.577
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr10_+_63331018
|
5.871
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr6_+_12398514
|
5.424
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr3_-_116272911
|
5.370
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr11_+_112337199
|
5.278
|
NM_000615
NM_001076682
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr6_+_114285209
|
4.901
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr1_-_191422346
|
4.701
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr4_-_187114407
|
4.588
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_+_70517863
|
4.380
|
|
SRGN
|
serglycin
|
|
chr10_+_70517823
|
4.219
|
NM_002727
|
SRGN
|
serglycin
|
|
chr8_+_70567581
|
4.214
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr1_+_85819004
|
4.118
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr7_-_27186400
|
3.938
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr8_+_70567634
|
3.899
|
|
SULF1
|
sulfatase 1
|
|
chr13_-_109757442
|
3.836
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr5_-_121441910
|
3.812
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr7_-_47588160
|
3.628
|
|
TNS3
|
tensin 3
|
|
chr7_-_47588266
|
3.559
|
|
TNS3
|
tensin 3
|
|
chr3_-_115825742
|
3.528
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_-_115960807
|
3.521
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chrX_-_99778340
|
3.502
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr4_-_186814852
|
3.443
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_111777651
|
3.384
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr1_-_85816520
|
3.360
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr1_-_85703198
|
3.320
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_-_175578156
|
3.319
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr4_-_186969041
|
3.275
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_-_215967442
|
3.159
|
|
FN1
|
fibronectin 1
|
|
chr6_+_148705421
|
3.106
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr7_-_27171673
|
3.028
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chrX_+_135079461
|
2.949
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr14_-_88953059
|
2.807
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr4_+_146623405
|
2.800
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr4_+_78297356
|
2.785
|
NM_004354
|
CCNG2
|
cyclin G2
|
|
chr6_-_128883125
|
2.755
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr6_+_89847019
|
2.689
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr2_-_152299229
|
2.647
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chrX_+_135079120
|
2.642
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_+_89847183
|
2.627
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr21_-_27261276
|
2.620
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr11_+_19328846
|
2.618
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chrX_-_13745108
|
2.611
|
|
GPM6B
|
glycoprotein M6B
|
|
chr7_-_47588652
|
2.515
|
|
TNS3
|
tensin 3
|
|
chr6_-_11340887
|
2.514
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr5_-_39461054
|
2.510
|
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr4_-_186969202
|
2.505
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_-_13744933
|
2.498
|
|
GPM6B
|
glycoprotein M6B
|
|
chr1_+_81544432
|
2.490
|
|
LPHN2
|
latrophilin 2
|
|
chr5_-_39460687
|
2.437
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr7_+_134226690
|
2.432
|
NM_033139
NM_033140
|
CALD1
|
caldesmon 1
|
|
chr2_+_189547377
|
2.428
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr7_+_93388951
|
2.416
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr18_+_71051713
|
2.403
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr5_+_15553288
|
2.361
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr2_-_190635566
|
2.360
|
NM_005259
|
MSTN
|
myostatin
|
|
chr4_-_111777584
|
2.347
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr2_-_162808123
|
2.326
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr12_+_13240987
|
2.316
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr7_+_134114912
|
2.306
|
|
CALD1
|
caldesmon 1
|
|
chr4_-_101658094
|
2.223
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr9_-_14303899
|
2.187
|
|
NFIB
|
nuclear factor I/B
|
|
chr9_-_14304036
|
2.170
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr8_-_18710575
|
2.168
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr4_-_186815116
|
2.167
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_16989371
|
2.144
|
NM_012334
|
MYO10
|
myosin X
|
|
chr12_-_90029672
|
2.142
|
NM_002345
|
LUM
|
lumican
|
|
chr2_+_69093641
|
2.134
|
NM_018153
NM_032208
NM_053034
|
ANTXR1
|
anthrax toxin receptor 1
|
|
chr7_-_47545717
|
2.133
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr1_-_199742871
|
2.122
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr1_-_95165089
|
2.121
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr6_-_11340869
|
2.105
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr7_+_134114701
|
2.082
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr5_-_111340421
|
2.058
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_-_101077605
|
2.048
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr5_-_121441805
|
2.041
|
|
LOX
|
lysyl oxidase
|
|
chr9_+_115382760
|
2.037
|
NM_021106
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr4_+_78297418
|
2.036
|
|
CCNG2
|
cyclin G2
|
|
chr12_-_90029328
|
2.022
|
|
LUM
|
lumican
|
|
chr5_-_121441866
|
2.008
|
|
LOX
|
lysyl oxidase
|
|
chr5_-_168204430
|
2.006
|
|
|
|
|
chr6_+_148705646
|
2.004
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr9_+_115382990
|
2.004
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_-_199742841
|
1.998
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr7_-_27171648
|
1.978
|
|
HOXA9
|
homeobox A9
|
|
chr7_+_94135384
|
1.962
|
|
PEG10
|
paternally expressed 10
|
|
chr1_+_242283129
|
1.902
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr17_-_59817654
|
1.893
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr13_-_32678142
|
1.883
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr6_+_148705817
|
1.878
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr9_+_36026900
|
1.828
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr1_-_199742864
|
1.826
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr3_-_55496607
|
1.813
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr17_-_59817456
|
1.788
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr7_-_27190882
|
1.783
|
|
HOXA11
|
homeobox A11
|
|
chr9_+_115303527
|
1.782
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr3_+_156280338
|
1.777
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr7_+_134226727
|
1.765
|
|
CALD1
|
caldesmon 1
|
|
chr3_+_156280129
|
1.764
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr2_+_36436873
|
1.751
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr5_+_86599350
|
1.748
|
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr3_-_150903749
|
1.713
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr17_-_59817722
|
1.683
|
NM_000442
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr5_-_39460787
|
1.664
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr1_-_199742589
|
1.657
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr5_-_83052606
|
1.652
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr2_-_188021117
|
1.644
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr7_+_115952279
|
1.640
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr3_-_55496370
|
1.632
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr2_-_151052391
|
1.626
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr12_-_16649211
|
1.622
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr9_-_14303509
|
1.621
|
|
NFIB
|
nuclear factor I/B
|
|
chr11_+_20001130
|
1.616
|
NM_001111019
|
NAV2
|
neuron navigator 2
|
|
chr7_-_111633620
|
1.613
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr14_-_51605487
|
1.606
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr4_+_91035025
|
1.600
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr1_-_56817823
|
1.599
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr1_+_99884018
|
1.585
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr3_-_151171576
|
1.581
|
|
PFN2
|
profilin 2
|
|
chr3_-_101316038
|
1.568
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr12_-_6103945
|
1.562
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr1_+_154120170
|
1.557
|
|
SYT11
|
synaptotagmin XI
|
|
chr2_-_210876560
|
1.554
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr9_-_23816062
|
1.531
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr2_-_86418143
|
1.519
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr7_+_115952325
|
1.512
|
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr10_-_91092410
|
1.511
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chrX_-_100801472
|
1.497
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr7_-_107430566
|
1.497
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr17_-_59817575
|
1.487
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr16_+_31391998
|
1.483
|
NM_015927
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr3_+_160269734
|
1.467
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr4_-_40631397
|
1.466
|
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr17_-_10393664
|
1.452
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr15_+_22751162
|
1.442
|
NM_003097
NM_022804
NM_005678
|
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N
SNRPN upstream reading frame
|
|
chr3_-_147361912
|
1.429
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr16_+_64958025
|
1.406
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr14_-_51605461
|
1.406
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr9_-_74735669
|
1.405
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr1_+_149299490
|
1.401
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr3_-_124822113
|
1.400
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chrX_-_13745234
|
1.399
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr1_-_95165037
|
1.384
|
|
CNN3
|
calponin 3, acidic
|
|
chr14_-_91483325
|
1.383
|
|
FBLN5
|
fibulin 5
|
|
chrX_+_114701432
|
1.378
|
NM_005032
|
PLS3
|
plastin 3
|
|
chr12_+_13240868
|
1.374
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chrX_-_10605726
|
1.363
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr16_+_54072969
|
1.362
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr2_-_39517633
|
1.362
|
NM_003618
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr1_-_56817568
|
1.361
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_+_101874765
|
1.361
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr2_+_101874809
|
1.360
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr9_-_16717887
|
1.358
|
|
BNC2
|
basonuclin 2
|
|
chr14_-_91483757
|
1.356
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr21_+_34112456
|
1.354
|
|
ITSN1
|
intersectin 1 (SH3 domain protein)
|
|
chr14_-_51605515
|
1.353
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr4_+_40953652
|
1.349
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr7_+_94130752
|
1.344
|
|
PEG10
|
paternally expressed 10
|
|
chr1_+_161305188
|
1.339
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr3_+_62280435
|
1.335
|
NM_020685
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr7_-_130891861
|
1.334
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr4_+_40953666
|
1.331
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr5_-_39430180
|
1.329
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr18_-_51406359
|
1.324
|
|
TCF4
|
transcription factor 4
|
|
chr11_+_112337299
|
1.322
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr11_+_112337411
|
1.319
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr6_-_134540666
|
1.319
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_+_140719773
|
1.318
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr1_+_84402962
|
1.315
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr9_-_36390817
|
1.314
|
NM_194328
NM_194332
|
RNF38
|
ring finger protein 38
|
|
chr12_+_69046241
|
1.311
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr3_-_124821868
|
1.309
|
|
MYLK
|
myosin light chain kinase
|
|
chr22_+_36422940
|
1.305
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr5_+_135392482
|
1.301
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr3_-_188936941
|
1.300
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr13_-_32658215
|
1.300
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_-_218551786
|
1.294
|
|
TNS1
|
tensin 1
|
|
chr5_-_111120817
|
1.294
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr10_+_31650069
|
1.290
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr4_+_41395493
|
1.288
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr15_-_21483457
|
1.287
|
|
NDN
|
necdin homolog (mouse)
|
|
chr20_+_19818209
|
1.285
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr12_-_109842736
|
1.281
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chr4_-_111777956
|
1.280
|
NM_153426
NM_153427
|
PITX2
|
paired-like homeodomain 2
|
|
chr9_-_36390212
|
1.279
|
NM_022781
NM_194329
|
RNF38
|
ring finger protein 38
|
|
chr1_-_56817802
|
1.278
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chrX_+_51944658
|
1.277
|
NM_001098800
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr3_+_189425886
|
1.277
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr17_-_45617899
|
1.268
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr1_-_56817717
|
1.267
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr17_+_16886491
|
1.258
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chrX_+_55951280
|
1.255
|
|
|
|
|
chrX_+_100220491
|
1.236
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr8_-_82557956
|
1.236
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr3_+_35697432
|
1.227
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr13_+_97593469
|
1.219
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr22_+_36928972
|
1.214
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr15_-_53996597
|
1.211
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr17_-_10265991
|
1.209
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|