|
chr16_-_87535106
|
2.510
|
NM_175931
|
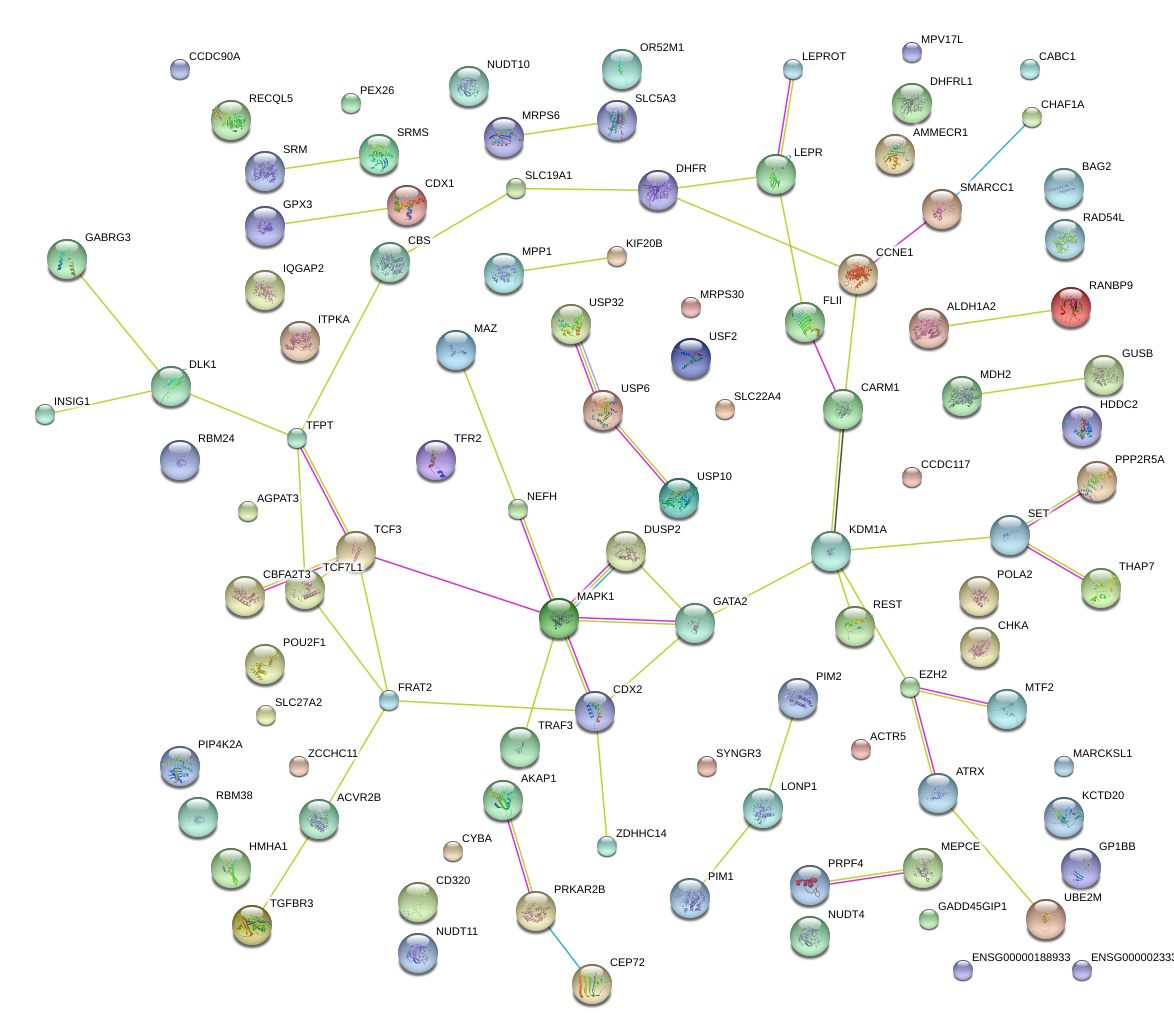
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr16_+_29725311
|
2.112
|
NM_001042539
NM_002383
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr21_-_45786709
|
1.855
|
NM_194255
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1
|
|
chr15_-_56145140
|
1.784
|
NM_003888
NM_170696
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr1_-_32574185
|
1.731
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr7_+_106472243
|
1.718
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr5_+_665404
|
1.615
|
NM_018140
|
CEP72
|
centrosomal protein 72kDa
|
|
chr5_+_665454
|
1.601
|
|
CEP72
|
centrosomal protein 72kDa
|
|
chr7_+_154720547
|
1.591
|
|
INSIG1
|
insulin induced gene 1
|
|
chr3_-_47798279
|
1.557
|
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr15_+_39573363
|
1.500
|
NM_002220
|
ITPKA
|
inositol 1,4,5-trisphosphate 3-kinase A
|
|
chr15_+_48261649
|
1.493
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr20_+_55399847
|
1.467
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr3_-_47798341
|
1.455
|
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr3_-_47798308
|
1.454
|
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr3_-_47798347
|
1.449
|
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr6_+_37245860
|
1.416
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr12_+_92295738
|
1.414
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr16_+_15397120
|
1.407
|
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr3_-_47798337
|
1.405
|
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr4_+_57468847
|
1.404
|
|
REST
|
RE1-silencing transcription factor
|
|
chr21_+_34367678
|
1.390
|
NM_032476
NM_006933
|
MRPS6
SLC5A3
|
mitochondrial ribosomal protein S6
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3
|
|
chr5_+_75734759
|
1.385
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr1_-_92124064
|
1.384
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr19_-_1603325
|
1.362
|
NM_003200
|
TCF3
|
transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47)
|
|
chr7_-_148212312
|
1.354
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr6_+_57145292
|
1.353
|
|
BAG2
|
BCL2-associated athanogene 2
|
|
chr3_-_47798381
|
1.350
|
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr7_-_148212230
|
1.308
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr22_+_18091065
|
1.308
|
NM_000407
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide
|
|
chr14_+_100262916
|
1.291
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr2_-_96174865
|
1.291
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr9_+_130491340
|
1.281
|
|
SET
|
SET nuclear oncogene
|
|
chr19_+_34994701
|
1.249
|
NM_001238
|
CCNE1
|
cyclin E1
|
|
chr5_+_131657964
|
1.245
|
NM_003059
|
SLC22A4
|
solute carrier family 22 (organic cation/ergothioneine transporter), member 4
|
|
chr3_-_47798372
|
1.235
|
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr7_-_100068612
|
1.233
|
|
TFR2
|
transferrin receptor 2
|
|
chr6_-_13819463
|
1.231
|
|
RANBP9
|
RAN binding protein 9
|
|
chr7_-_148212310
|
1.227
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr7_-_148212336
|
1.224
|
NM_004456
NM_152998
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr7_+_99865189
|
1.209
|
NM_019606
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr6_-_13819931
|
1.183
|
|
RANBP9
|
RAN binding protein 9
|
|
chrX_-_153686923
|
1.182
|
NM_001166460
NM_001166461
NM_001166462
NM_002436
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa
|
|
chr19_+_1022208
|
1.181
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr10_-_99084417
|
1.177
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr6_+_157722467
|
1.155
|
NM_024630
NM_153746
|
ZDHHC14
|
zinc finger, DHHC-type containing 14
|
|
chr16_+_15397087
|
1.141
|
NM_001128423
NM_173803
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr21_-_43368963
|
1.139
|
NM_000071
|
CBS
|
cystathionine-beta-synthase
|
|
chr1_+_46485972
|
1.123
|
|
RAD54L
|
RAD54-like (S. cerevisiae)
|
|
chr7_+_99865340
|
1.118
|
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr14_+_100262943
|
1.108
|
NM_003836
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr1_-_52791309
|
1.105
|
NM_001009881
NM_015269
|
ZCCHC11
|
zinc finger, CCHC domain containing 11
|
|
chr5_+_149526533
|
1.103
|
NM_001804
|
CDX1
|
caudal type homeobox 1
|
|
chr1_+_93317374
|
1.097
|
NM_001164391
NM_001164392
NM_001164393
NM_007358
|
MTF2
|
metal response element binding transcription factor 2
|
|
chr3_-_47798390
|
1.079
|
NM_003074
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr15_+_24799174
|
1.076
|
NM_033223
|
GABRG3
|
gamma-aminobutyric acid (GABA) A receptor, gamma 3
|
|
chr6_-_13922525
|
1.067
|
NM_001031713
|
CCDC90A
|
coiled-coil domain containing 90A
|
|
chr19_+_40451626
|
1.058
|
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chr22_+_27498679
|
1.052
|
|
CCDC117
|
coiled-coil domain containing 117
|
|
chr16_-_87244896
|
1.051
|
NM_000101
|
CYBA
|
cytochrome b-245, alpha polypeptide
|
|
chr1_-_11042668
|
1.044
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr14_+_100263028
|
1.038
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr16_+_83291097
|
1.033
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr16_+_83291112
|
1.025
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr7_-_65084668
|
1.024
|
NM_000181
|
GUSB
|
glucuronidase, beta
|
|
chr1_+_225194582
|
1.017
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr16_+_83291055
|
1.016
|
NM_005153
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr6_-_13819622
|
1.009
|
NM_005493
|
RANBP9
|
RAN binding protein 9
|
|
chr1_+_46485946
|
0.997
|
NM_001142548
NM_003579
|
RAD54L
|
RAD54-like (S. cerevisiae)
|
|
chr22_-_19686351
|
0.988
|
NM_001008695
NM_030573
|
THAP7
|
THAP domain containing 7
|
|
chr1_+_225194622
|
0.986
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr11_-_67645246
|
0.986
|
|
CHKA
|
choline kinase alpha
|
|
chr10_-_23043449
|
0.985
|
NM_005028
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr17_+_26060617
|
0.978
|
|
|
|
|
chr3_+_38470782
|
0.978
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr20_+_36810510
|
0.970
|
NM_024855
|
ACTR5
|
ARP5 actin-related protein 5 homolog (yeast)
|
|
chr16_+_2593351
|
0.966
|
|
LOC652276
|
hypothetical LOC652276
|
|
chr3_-_129690056
|
0.964
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr1_+_23218515
|
0.962
|
NM_001009999
NM_015013
|
KDM1A
|
lysine (K)-specific demethylase 1A
|
|
chr19_-_8279157
|
0.959
|
NM_001165895
NM_016579
|
CD320
|
CD320 molecule
|
|
chr16_+_29725597
|
0.947
|
|
|
|
|
chrX_-_109447970
|
0.946
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr16_+_1979959
|
0.945
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr9_+_115077752
|
0.941
|
|
PRPF4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast)
|
|
chr14_+_102313585
|
0.938
|
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr1_+_165456572
|
0.936
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr22_+_16940755
|
0.932
|
|
PEX26
|
peroxisomal biogenesis factor 26
|
|
chr1_+_225194560
|
0.931
|
NM_020247
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr6_-_125664913
|
0.900
|
NM_016063
|
HDDC2
|
HD domain containing 2
|
|
chr5_-_79986539
|
0.895
|
NM_000791
|
DHFR
|
dihydrofolate reductase
|
|
chr21_+_44109454
|
0.889
|
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr19_+_10843244
|
0.888
|
NM_199141
|
CARM1
|
coactivator-associated arginine methyltransferase 1
|
|
chr1_+_65763945
|
0.883
|
NM_001198687
NM_001198688
NM_001198689
|
LEPR
|
leptin receptor
|
|
chr6_+_36518778
|
0.880
|
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr1_+_225194616
|
0.880
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chrX_-_48661226
|
0.874
|
NM_006875
|
PIM2
|
pim-2 oncogene
|
|
chr7_+_154720416
|
0.872
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr1_+_210525490
|
0.870
|
NM_006243
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr5_+_44844724
|
0.870
|
NM_016640
|
MRPS30
|
mitochondrial ribosomal protein S30
|
|
chr1_+_23218572
|
0.869
|
|
KDM1A
|
lysine (K)-specific demethylase 1A
|
|
chr7_+_75515338
|
0.865
|
|
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial)
|
|
chrX_-_153686818
|
0.855
|
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa
|
|
chr1_+_65763978
|
0.854
|
|
LEPR
|
leptin receptor
|
|
chr16_-_34262262
|
0.853
|
|
UBE2MP1
|
ubiquitin-conjugating enzyme E2M pseudogene 1
|
|
chr22_-_20551937
|
0.853
|
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr17_+_52517529
|
0.842
|
NM_003488
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr11_+_64785888
|
0.839
|
NM_002689
|
POLA2
|
polymerase (DNA directed), alpha 2 (70kD subunit)
|
|
chr19_+_4353658
|
0.834
|
NM_005483
|
CHAF1A
|
chromatin assembly factor 1, subunit A (p150)
|
|
chr19_-_12929008
|
0.833
|
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1
|
|
chr22_+_28206180
|
0.832
|
NM_021076
|
NEFH
|
neurofilament, heavy polypeptide
|
|
chr19_-_16443810
|
0.827
|
|
EPS15L1
|
epidermal growth factor receptor pathway substrate 15-like 1
|
|
chr7_+_87343479
|
0.824
|
NM_006716
|
DBF4
|
DBF4 homolog (S. cerevisiae)
|
|
chr16_+_84204424
|
0.821
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr14_+_100263066
|
0.817
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr16_-_28542366
|
0.810
|
NM_177536
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr3_-_50515857
|
0.810
|
NM_001005505
NM_001174051
NM_006030
|
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2
|
|
chr6_+_36518521
|
0.808
|
NM_173562
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr19_+_40451720
|
0.807
|
NM_003367
NM_207291
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chr1_+_11674365
|
0.804
|
NM_198545
|
C1orf187
|
chromosome 1 open reading frame 187
|
|
chr7_+_149696792
|
0.799
|
NM_001099695
NM_001099696
NM_013400
|
REPIN1
|
replication initiator 1
|
|
chr21_+_29593098
|
0.799
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr4_+_57469122
|
0.796
|
|
REST
|
RE1-silencing transcription factor
|
|
chr15_+_83092806
|
0.796
|
NM_014630
|
ZNF592
|
zinc finger protein 592
|
|
chr22_+_38183262
|
0.787
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr12_+_2938703
|
0.784
|
NM_003213
NM_201441
NM_201443
|
TEAD4
|
TEA domain family member 4
|
|
chr9_-_100510657
|
0.783
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr9_-_139100923
|
0.780
|
|
LOC100289341
|
similar to hCG2022304
|
|
chr18_-_12367248
|
0.777
|
NM_006796
|
AFG3L2
|
AFG3 ATPase family gene 3-like 2 (S. cerevisiae)
|
|
chr16_+_79598016
|
0.776
|
|
CENPN
|
centromere protein N
|
|
chr22_+_20101661
|
0.770
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chrX_-_109447954
|
0.768
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr7_-_87343599
|
0.764
|
NM_018843
|
SLC25A40
|
solute carrier family 25, member 40
|
|
chr9_+_130491370
|
0.762
|
|
SET
|
SET nuclear oncogene
|
|
chr12_-_116021473
|
0.760
|
NM_001168325
NM_017899
|
TESC
|
tescalcin
|
|
chr1_+_32889335
|
0.759
|
NM_001135255
NM_005610
NM_001135256
|
RBBP4
|
retinoblastoma binding protein 4
|
|
chr1_-_218168548
|
0.758
|
NM_018713
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr5_+_118719448
|
0.757
|
NM_014350
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr5_+_70918888
|
0.757
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr2_-_27288518
|
0.754
|
NM_021095
|
SLC5A6
|
solute carrier family 5 (sodium-dependent vitamin transporter), member 6
|
|
chr16_-_4341266
|
0.753
|
NM_016069
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae)
|
|
chr12_+_6893751
|
0.752
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr12_-_107479082
|
0.749
|
|
SART3
|
squamous cell carcinoma antigen recognized by T cells 3
|
|
chr10_-_23043042
|
0.748
|
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr7_-_148212292
|
0.747
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr16_+_83291107
|
0.747
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr11_+_67836575
|
0.745
|
NM_002335
|
LRP5
|
low density lipoprotein receptor-related protein 5
|
|
chr8_+_56954953
|
0.744
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr5_+_70918935
|
0.743
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr6_-_43305156
|
0.741
|
NM_006443
NM_199184
|
C6orf108
|
chromosome 6 open reading frame 108
|
|
chr18_+_2645885
|
0.738
|
NM_015295
|
SMCHD1
|
structural maintenance of chromosomes flexible hinge domain containing 1
|
|
chr5_+_70918893
|
0.738
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr12_-_107479072
|
0.732
|
|
SART3
|
squamous cell carcinoma antigen recognized by T cells 3
|
|
chr22_+_18081968
|
0.731
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr6_+_34312906
|
0.728
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_-_159413833
|
0.723
|
NM_003779
|
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3
|
|
chr7_-_150305933
|
0.721
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr12_-_107479059
|
0.719
|
|
SART3
|
squamous cell carcinoma antigen recognized by T cells 3
|
|
chr8_+_61753793
|
0.718
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr1_-_53566331
|
0.717
|
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chrX_-_137621492
|
0.716
|
NM_004114
|
FGF13
|
fibroblast growth factor 13
|
|
chr1_-_39111422
|
0.713
|
NM_012333
|
MYCBP
|
c-myc binding protein
|
|
chr7_+_99613282
|
0.711
|
|
STAG3
|
stromal antigen 3
|
|
chr10_+_30762862
|
0.710
|
NM_005204
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr19_-_12929033
|
0.709
|
NM_052850
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1
|
|
chr17_+_70769539
|
0.708
|
|
MRPS7
|
mitochondrial ribosomal protein S7
|
|
chr1_-_11042634
|
0.707
|
|
SRM
|
spermidine synthase
|
|
chr7_-_23476156
|
0.705
|
|
|
|
|
chr2_-_203444468
|
0.705
|
NM_138468
|
ICA1L
|
islet cell autoantigen 1,69kDa-like
|
|
chr10_+_28861705
|
0.704
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr17_-_3814334
|
0.702
|
NM_005173
NM_174953
NM_174954
NM_174955
NM_174956
NM_174957
NM_174958
|
ATP2A3
|
ATPase, Ca++ transporting, ubiquitous
|
|
chr12_-_6586811
|
0.700
|
NM_001273
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr4_-_185892204
|
0.699
|
NM_024629
|
MLF1IP
|
MLF1 interacting protein
|
|
chr1_+_91739241
|
0.698
|
NM_001134420
|
CDC7
|
cell division cycle 7 homolog (S. cerevisiae)
|
|
chr19_-_10474767
|
0.695
|
NM_203500
|
KEAP1
|
kelch-like ECH-associated protein 1
|
|
chr10_+_22650145
|
0.695
|
|
BMI1
|
BMI1 polycomb ring finger oncogene
|
|
chr6_+_89912464
|
0.695
|
NM_001010853
|
PM20D2
|
peptidase M20 domain containing 2
|
|
chr19_+_10843369
|
0.694
|
|
CARM1
|
coactivator-associated arginine methyltransferase 1
|
|
chr3_+_73128771
|
0.691
|
NM_174907
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr17_+_70769600
|
0.689
|
|
MRPS7
|
mitochondrial ribosomal protein S7
|
|
chr16_-_3707450
|
0.686
|
NM_016292
|
TRAP1
|
TNF receptor-associated protein 1
|
|
chr21_+_44109529
|
0.685
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr11_-_67645433
|
0.685
|
NM_001277
NM_212469
|
CHKA
|
choline kinase alpha
|
|
chr18_-_12367168
|
0.684
|
|
AFG3L2
|
AFG3 ATPase family gene 3-like 2 (S. cerevisiae)
|
|
chr6_+_96132064
|
0.683
|
NM_024641
|
MANEA
|
mannosidase, endo-alpha
|
|
chr6_-_125664770
|
0.683
|
|
HDDC2
|
HD domain containing 2
|
|
chr3_+_154035425
|
0.680
|
NM_002563
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1
|
|
chr4_-_2934945
|
0.679
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr6_+_135544138
|
0.678
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr1_+_91738953
|
0.677
|
NM_001134419
NM_003503
|
CDC7
|
cell division cycle 7 homolog (S. cerevisiae)
|
|
chr2_-_10506033
|
0.675
|
|
ODC1
|
ornithine decarboxylase 1
|
|
chr17_-_73636386
|
0.667
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr12_-_6586736
|
0.665
|
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr19_+_54314292
|
0.665
|
NM_003660
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr3_-_13436802
|
0.663
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr10_-_28861465
|
0.662
|
|
LOC220906
|
hypothetical LOC220906
|
|
chr7_-_99536214
|
0.662
|
NM_182776
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr1_+_181075104
|
0.662
|
|
DHX9
|
DEAH (Asp-Glu-Ala-His) box polypeptide 9
|
|
chr9_+_130491291
|
0.661
|
NM_003011
|
SET
|
SET nuclear oncogene
|
|
chr5_+_70918856
|
0.661
|
NM_022132
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr8_-_145609737
|
0.661
|
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|