|
chr6_+_135544214
|
3.884
|
|
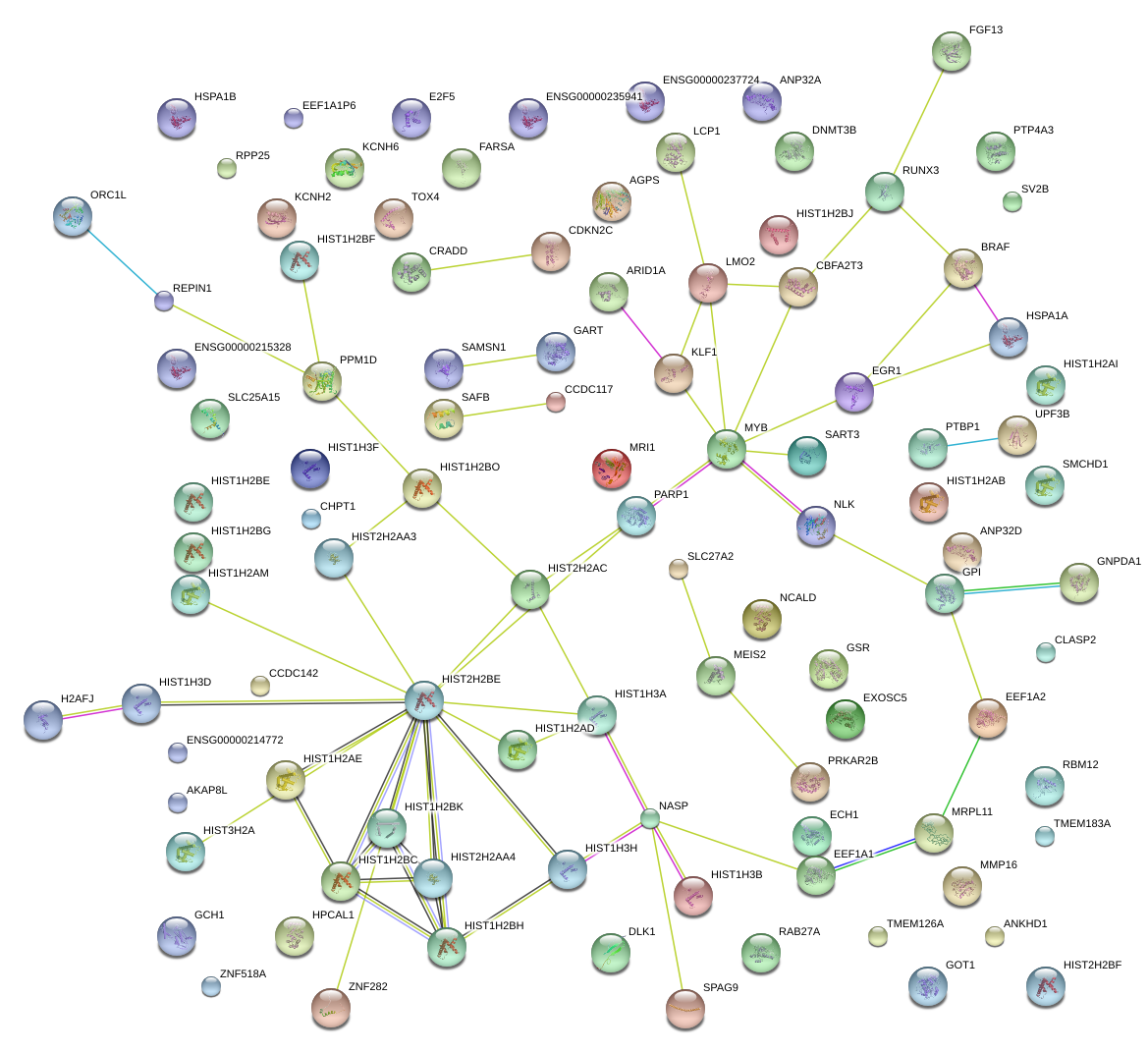
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr19_-_12858956
|
3.698
|
NM_006563
|
KLF1
|
Kruppel-like factor 1 (erythroid)
|
|
chr17_+_23393744
|
3.441
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr1_+_148089251
|
3.273
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr1_-_148080888
|
2.970
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr6_+_135544170
|
2.806
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr6_+_135544138
|
2.767
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr2_+_177965734
|
2.726
|
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr15_+_48261649
|
2.713
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr7_+_148523486
|
2.666
|
NM_003575
|
ZNF282
|
zinc finger protein 282
|
|
chr2_+_177965617
|
2.632
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr20_-_33706532
|
2.622
|
|
RBM12
|
RNA binding motif protein 12
|
|
chr19_-_12905528
|
2.522
|
NM_004461
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chrX_-_137621060
|
2.519
|
|
FGF13
|
fibroblast growth factor 13
|
|
chr6_-_26324850
|
2.401
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr14_+_100270222
|
2.390
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr19_-_12905524
|
2.390
|
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr15_-_73035978
|
2.326
|
|
RPP25
|
ribonuclease P/MRP 25kDa subunit
|
|
chr19_-_46595053
|
2.226
|
NM_020158
|
EXOSC5
|
exosome component 5
|
|
chr11_-_33847834
|
2.156
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr1_+_148125090
|
2.125
|
NM_003517
|
HIST2H2AC
|
histone cluster 2, H2ac
|
|
chr19_-_12905459
|
2.105
|
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr7_-_140270749
|
2.103
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr11_-_33847937
|
2.101
|
NM_001142316
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr6_+_26291936
|
2.065
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr19_+_5574119
|
1.996
|
|
SAFB
|
scaffold attachment factor B
|
|
chr14_-_54438913
|
1.958
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr10_+_97879457
|
1.945
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr21_-_14840506
|
1.936
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr6_+_27883877
|
1.916
|
NM_003509
|
HIST1H2AI
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H3f
|
|
chr8_-_30704762
|
1.907
|
|
GSR
|
glutathione reductase
|
|
chr1_-_148050534
|
1.899
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr15_-_73036179
|
1.881
|
|
RPP25
|
ribonuclease P/MRP 25kDa subunit
|
|
chr6_-_74284522
|
1.858
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr12_+_100615530
|
1.840
|
NM_020244
|
CHPT1
|
choline phosphotransferase 1
|
|
chr6_+_27885692
|
1.818
|
NM_003536
|
HIST1H3H
|
histone cluster 1, H3h
|
|
chr7_-_150283794
|
1.776
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr1_+_45822246
|
1.775
|
NM_001195193
NM_002482
NM_152298
|
NASP
|
nuclear autoantigenic sperm protein (histone-binding)
|
|
chr21_-_33836994
|
1.762
|
NM_001136005
NM_001136006
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase
|
|
chr1_+_51208499
|
1.726
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr2_-_74563864
|
1.724
|
NM_032779
|
CCDC142
|
coiled-coil domain containing 142
|
|
chr8_-_30704984
|
1.712
|
NM_000637
NM_001195102
NM_001195103
NM_001195104
|
GSR
|
glutathione reductase
|
|
chr15_+_89570333
|
1.700
|
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr19_+_13736345
|
1.681
|
|
MRI1
|
methylthioribose-1-phosphate isomerase homolog (S. cerevisiae)
|
|
chr17_-_46479127
|
1.677
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr1_+_26973804
|
1.674
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr15_-_35179185
|
1.665
|
|
MEIS2
|
Meis homeobox 2
|
|
chr11_-_33847507
|
1.663
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr6_-_27222556
|
1.645
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr1_-_25164061
|
1.641
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr1_-_224662156
|
1.603
|
|
PARP1
|
poly (ADP-ribose) polymerase 1
|
|
chr7_+_106472243
|
1.574
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr5_+_137829065
|
1.570
|
|
EGR1
|
early growth response 1
|
|
chr19_+_748444
|
1.564
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr15_-_66900266
|
1.562
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chr6_-_26305456
|
1.527
|
|
HIST1H3D
|
histone cluster 1, H3d
|
|
chr6_+_27208763
|
1.514
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr6_+_26307691
|
1.509
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr8_+_86276893
|
1.499
|
|
|
|
|
chr22_+_27498679
|
1.490
|
|
CCDC117
|
coiled-coil domain containing 117
|
|
chr19_-_44014228
|
1.473
|
NM_001398
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal
|
|
chr11_-_65962836
|
1.471
|
NM_016050
NM_170738
NM_170739
|
MRPL11
|
mitochondrial ribosomal protein L11
|
|
chr8_+_86276870
|
1.461
|
NM_001083588
NM_001951
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr6_+_26325124
|
1.450
|
NM_021052
|
HIST1H2AE
|
histone cluster 1, H2ae
|
|
chr8_+_142501188
|
1.436
|
NM_007079
NM_032611
|
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3
|
|
chr19_-_15390816
|
1.424
|
NM_014371
|
AKAP8L
|
A kinase (PRKA) anchor protein 8-like
|
|
chr1_-_52642646
|
1.421
|
NM_001190818
NM_001190819
NM_004153
|
ORC1
|
origin recognition complex, subunit 1
|
|
chr19_+_13736352
|
1.418
|
|
MRI1
|
methylthioribose-1-phosphate isomerase homolog (S. cerevisiae)
|
|
chr1_+_201243136
|
1.417
|
|
TMEM183A
|
transmembrane protein 183A
|
|
chr15_-_53350383
|
1.415
|
NM_183236
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr7_+_149696901
|
1.410
|
|
REPIN1
|
replication initiator 1
|
|
chr20_+_30831298
|
1.402
|
NM_175850
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr12_+_92595281
|
1.400
|
NM_003805
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain
|
|
chr12_-_107479082
|
1.390
|
|
SART3
|
squamous cell carcinoma antigen recognized by T cells 3
|
|
chr13_-_45640679
|
1.385
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr6_-_26140266
|
1.381
|
NM_003537
|
HIST1H3B
|
histone cluster 1, H3b
|
|
chr12_-_107479072
|
1.376
|
|
SART3
|
squamous cell carcinoma antigen recognized by T cells 3
|
|
chr17_+_56032385
|
1.374
|
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr12_-_107479059
|
1.372
|
|
SART3
|
squamous cell carcinoma antigen recognized by T cells 3
|
|
chr1_-_226712140
|
1.358
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr6_+_31891509
|
1.356
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr3_-_33675484
|
1.334
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr5_+_139856688
|
1.331
|
|
ANKHD1
|
ankyrin repeat and KH domain containing 1
|
|
chrX_-_118870936
|
1.329
|
NM_023010
NM_080632
|
UPF3B
|
UPF3 regulator of nonsense transcripts homolog B (yeast)
|
|
chr16_-_87570716
|
1.322
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr17_-_37423196
|
1.317
|
NM_003315
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7
|
|
chr2_+_74610333
|
1.314
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr1_-_148126089
|
1.314
|
NM_175065
|
HIST2H2AB
HIST2H2BE
|
histone cluster 2, H2ab
histone cluster 2, H2be
|
|
chr6_+_33276580
|
1.311
|
NM_001077516
NM_006979
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr19_-_55104875
|
1.302
|
|
NUP62
|
nucleoporin 62kDa
|
|
chr1_+_35507197
|
1.297
|
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr1_-_148124738
|
1.288
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr15_-_35178888
|
1.285
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr8_+_23442331
|
1.285
|
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr6_-_39305203
|
1.275
|
NM_003740
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr1_+_51208229
|
1.269
|
NM_078626
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr2_-_27288518
|
1.257
|
NM_021095
|
SLC5A6
|
solute carrier family 5 (sodium-dependent vitamin transporter), member 6
|
|
chr12_-_48246488
|
1.254
|
NM_001012300
|
MCRS1
|
microspherule protein 1
|
|
chrX_-_152938528
|
1.247
|
NM_001025242
NM_001025243
NM_001569
|
IRAK1
|
interleukin-1 receptor-associated kinase 1
|
|
chr6_+_26212082
|
1.238
|
NM_003542
|
HIST1H4C
|
histone cluster 1, H4c
|
|
chr10_+_59815110
|
1.237
|
NM_003201
|
TFAM
|
transcription factor A, mitochondrial
|
|
chr1_-_147665783
|
1.233
|
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chrX_+_128741578
|
1.225
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr8_+_23442307
|
1.218
|
NM_016612
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr16_-_28765138
|
1.211
|
NM_003321
|
TUFM
|
Tu translation elongation factor, mitochondrial
|
|
chr11_+_75203884
|
1.204
|
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr17_-_2187427
|
1.202
|
NM_018128
|
TSR1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae)
|
|
chr10_+_14920143
|
1.201
|
NM_016299
|
HSPA14
|
heat shock 70kDa protein 14
|
|
chr3_-_140148671
|
1.193
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr3_-_130323033
|
1.187
|
|
RAB43
|
RAB43, member RAS oncogene family
|
|
chr4_-_83514160
|
1.186
|
NM_001003810
NM_002138
NM_031369
NM_031370
|
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa)
|
|
chr8_-_108579270
|
1.180
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr1_-_199259447
|
1.175
|
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr1_+_39229474
|
1.170
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr18_+_9904249
|
1.169
|
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr19_+_51542085
|
1.165
|
NM_006247
|
PPP5C
|
protein phosphatase 5, catalytic subunit
|
|
chr14_-_22468445
|
1.164
|
NM_001039619
NM_006109
|
PRMT5
|
protein arginine methyltransferase 5
|
|
chr2_+_74610395
|
1.161
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr3_+_47841478
|
1.154
|
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30
|
|
chr6_+_26381122
|
1.144
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr3_-_172660545
|
1.139
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr1_+_201243101
|
1.138
|
|
TMEM183B
|
transmembrane protein 183B
|
|
chr6_+_35373572
|
1.135
|
NM_022047
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr10_+_21863099
|
1.133
|
NM_001195627
NM_001195628
NM_001195630
NM_004641
|
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10
|
|
chr1_-_243093833
|
1.132
|
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr11_+_59979857
|
1.129
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr10_-_64246129
|
1.128
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr6_+_43592758
|
1.125
|
NM_004875
NM_203290
|
POLR1C
|
polymerase (RNA) I polypeptide C, 30kDa
|
|
chr2_+_161981138
|
1.113
|
|
TBR1
|
T-box, brain, 1
|
|
chr11_-_118405288
|
1.112
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr12_-_107479294
|
1.109
|
NM_014706
|
SART3
|
squamous cell carcinoma antigen recognized by T cells 3
|
|
chr22_+_21011653
|
1.106
|
|
|
|
|
chr5_-_178977441
|
1.103
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr7_-_129038701
|
1.103
|
|
|
|
|
chr17_-_72244231
|
1.101
|
|
|
|
|
chr17_-_38385511
|
1.100
|
NM_025267
|
AARSD1
|
alanyl-tRNA synthetase domain containing 1
|
|
chrX_-_152938456
|
1.098
|
|
IRAK1
|
interleukin-1 receptor-associated kinase 1
|
|
chr6_+_43592812
|
1.094
|
|
POLR1C
|
polymerase (RNA) I polypeptide C, 30kDa
|
|
chr6_+_32254108
|
1.093
|
NM_006913
|
RNF5
|
ring finger protein 5
|
|
chr1_+_201243150
|
1.090
|
NM_001079809
NM_138391
|
TMEM183B
TMEM183A
|
transmembrane protein 183B
transmembrane protein 183A
|
|
chr7_-_99536966
|
1.083
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chrX_+_128741562
|
1.080
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr6_-_144458326
|
1.077
|
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa
|
|
chr19_-_15390738
|
1.075
|
|
AKAP8L
|
A kinase (PRKA) anchor protein 8-like
|
|
chr7_+_149696792
|
1.075
|
NM_001099695
NM_001099696
NM_013400
|
REPIN1
|
replication initiator 1
|
|
chr8_+_42315132
|
1.074
|
NM_002690
|
POLB
|
polymerase (DNA directed), beta
|
|
chr3_-_122862447
|
1.072
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr7_+_116447527
|
1.069
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr12_+_94776836
|
1.061
|
NM_003095
|
SNRPF
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr11_-_65895640
|
1.061
|
NM_001532
|
SLC29A2
|
solute carrier family 29 (nucleoside transporters), member 2
|
|
chr7_-_99536929
|
1.057
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr2_+_68238490
|
1.055
|
NM_020143
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae)
|
|
chr7_+_116447499
|
1.046
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr6_+_33365386
|
1.045
|
NM_001185181
|
PFDN6
|
prefoldin subunit 6
|
|
chr11_-_62363551
|
1.028
|
|
WDR74
|
WD repeat domain 74
|
|
chr6_-_33365264
|
1.020
|
NM_001164267
NM_005452
|
WDR46
|
WD repeat domain 46
|
|
chr16_-_29532538
|
1.018
|
|
|
|
|
chr6_-_27968906
|
1.016
|
NM_003514
|
HIST1H2AM
|
histone cluster 1, H2am
|
|
chr6_-_144458446
|
1.015
|
NM_031287
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa
|
|
chr7_-_23476497
|
1.013
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr22_+_21116283
|
1.011
|
|
|
|
|
chr2_-_1905508
|
1.009
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr6_+_32254188
|
1.008
|
|
RNF5
|
ring finger protein 5
|
|
chr12_-_54938385
|
1.007
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr5_+_61637993
|
1.000
|
|
KIF2A
|
kinesin heavy chain member 2A
|
|
chr6_-_27883662
|
0.999
|
NM_003519
|
HIST1H2BL
|
histone cluster 1, H2bl
|
|
chr18_+_9904202
|
0.997
|
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr2_+_74610751
|
0.994
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr11_-_5204769
|
0.990
|
NM_000518
|
HBB
|
hemoglobin, beta
|
|
chr20_-_45418818
|
0.990
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr8_-_145485907
|
0.989
|
NM_015201
|
BOP1
|
block of proliferation 1
|
|
chr6_-_31973418
|
0.987
|
|
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2
|
|
chr3_-_187138342
|
0.986
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr7_-_36992221
|
0.983
|
NM_130442
|
ELMO1
|
engulfment and cell motility 1
|
|
chr19_+_10673105
|
0.983
|
NM_031209
|
QTRT1
|
queuine tRNA-ribosyltransferase 1
|
|
chr15_-_66900143
|
0.981
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chrX_-_118711088
|
0.980
|
|
SEPT6
|
septin 6
|
|
chr5_-_79986539
|
0.980
|
NM_000791
|
DHFR
|
dihydrofolate reductase
|
|
chr7_-_99536852
|
0.980
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr10_-_64246080
|
0.975
|
|
EGR2
|
early growth response 2
|
|
chr17_-_72045218
|
0.972
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr5_+_126140746
|
0.965
|
|
LMNB1
|
lamin B1
|
|
chrX_-_118623840
|
0.960
|
NM_001173487
NM_017544
|
NKRF
|
NFKB repressing factor
|
|
chr2_-_158008763
|
0.958
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr2_+_105320247
|
0.956
|
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr6_-_27208507
|
0.956
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr20_-_48980858
|
0.954
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr5_-_175748533
|
0.953
|
|
NOP16
|
NOP16 nucleolar protein homolog (yeast)
|
|
chr8_+_23442413
|
0.951
|
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr4_-_151990061
|
0.948
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr1_+_42896634
|
0.938
|
NM_006347
|
PPIH
|
peptidylprolyl isomerase H (cyclophilin H)
|
|
chr16_+_88314884
|
0.934
|
NM_152287
|
ZNF276
|
zinc finger protein 276
|
|
chr14_-_91046383
|
0.933
|
NM_032560
|
SMEK1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium)
|
|
chr6_+_35373632
|
0.930
|
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr1_+_51855343
|
0.930
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr13_-_40491405
|
0.929
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr19_+_6723801
|
0.927
|
|
|
|
|
chr1_+_178190493
|
0.926
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr2_+_74610494
|
0.925
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr12_+_103375116
|
0.921
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|