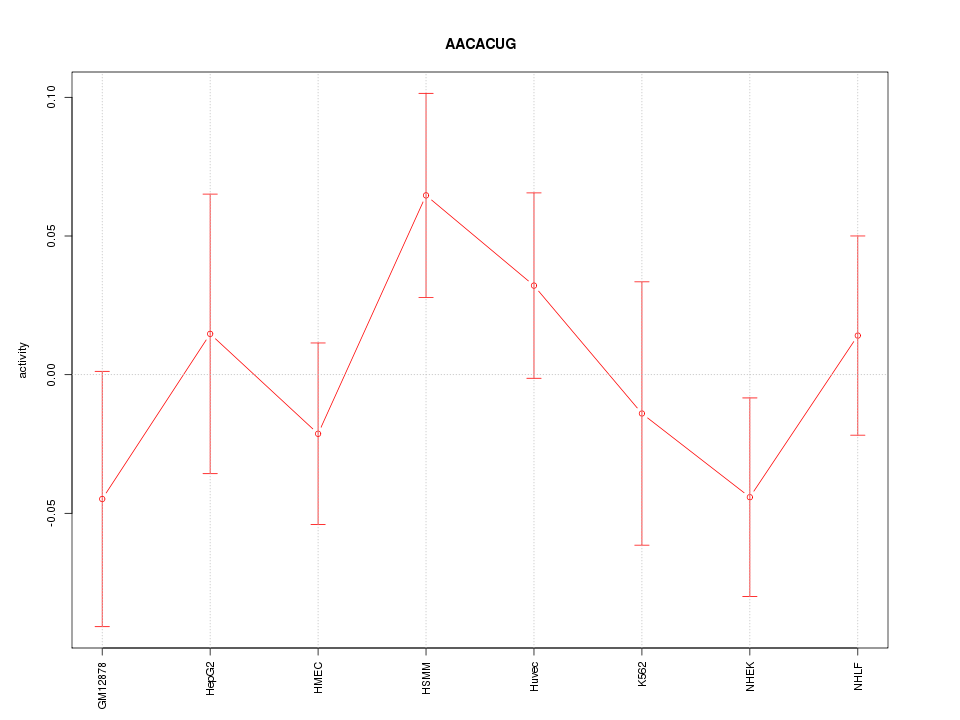
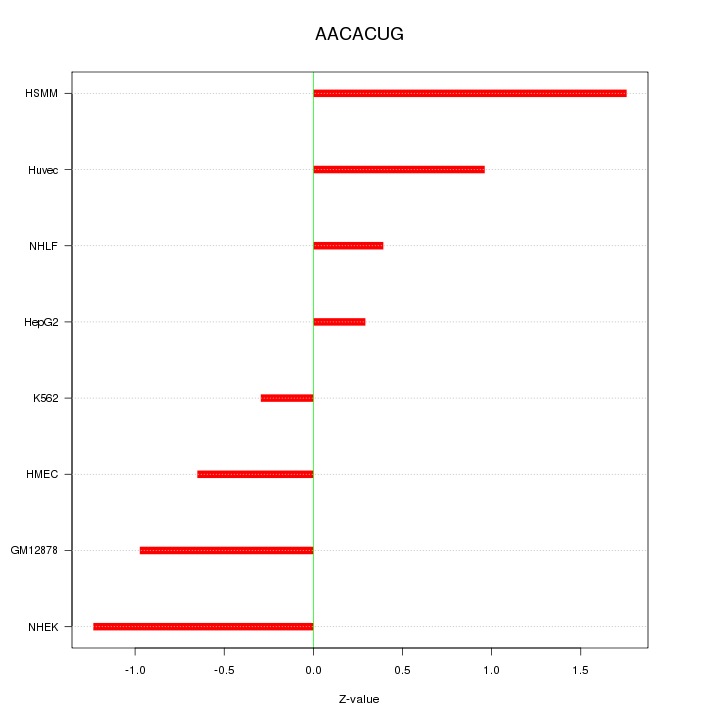
Motif ID: AACACUG
Z-value: 0.951

Mature miRNA associated with seed AACACUG:
| Name | miRBase Accession |
|---|---|
| hsa-miR-141-3p | MIMAT0000432 |
| hsa-miR-200a-3p | MIMAT0000682 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.2 | 1.6 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.1 | 0.4 | GO:0014028 | notochord formation(GO:0014028) axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.4 | GO:0061298 | progesterone secretion(GO:0042701) retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.4 | GO:0060340 | pericardium morphogenesis(GO:0003344) optic cup formation involved in camera-type eye development(GO:0003408) diencephalon morphogenesis(GO:0048852) positive regulation of type I interferon-mediated signaling pathway(GO:0060340) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.1 | 0.6 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.3 | GO:0045726 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.3 | GO:0032927 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of activin receptor signaling pathway(GO:0032927) positive regulation of follicle-stimulating hormone secretion(GO:0046881) Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.8 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.4 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.1 | 0.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.2 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 | 0.2 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 0.2 | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.1 | 0.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.1 | 0.5 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.2 | GO:0043276 | anoikis(GO:0043276) |
| 0.1 | 0.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0030812 | muscle hypertrophy in response to stress(GO:0003299) response to stimulus involved in regulation of muscle adaptation(GO:0014874) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 1.7 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 1.0 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.0 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.2 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.7 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.4 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.3 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.3 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.8 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:1902803 | axon target recognition(GO:0007412) synaptic vesicle priming(GO:0016082) regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 1.1 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.0 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.6 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.7 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.2 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0051589 | negative regulation of neurotransmitter transport(GO:0051589) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 4.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 1.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0034451 | exocyst(GO:0000145) centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.3 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.1 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST_GAQ_PATHWAY | G alpha q Pathway |