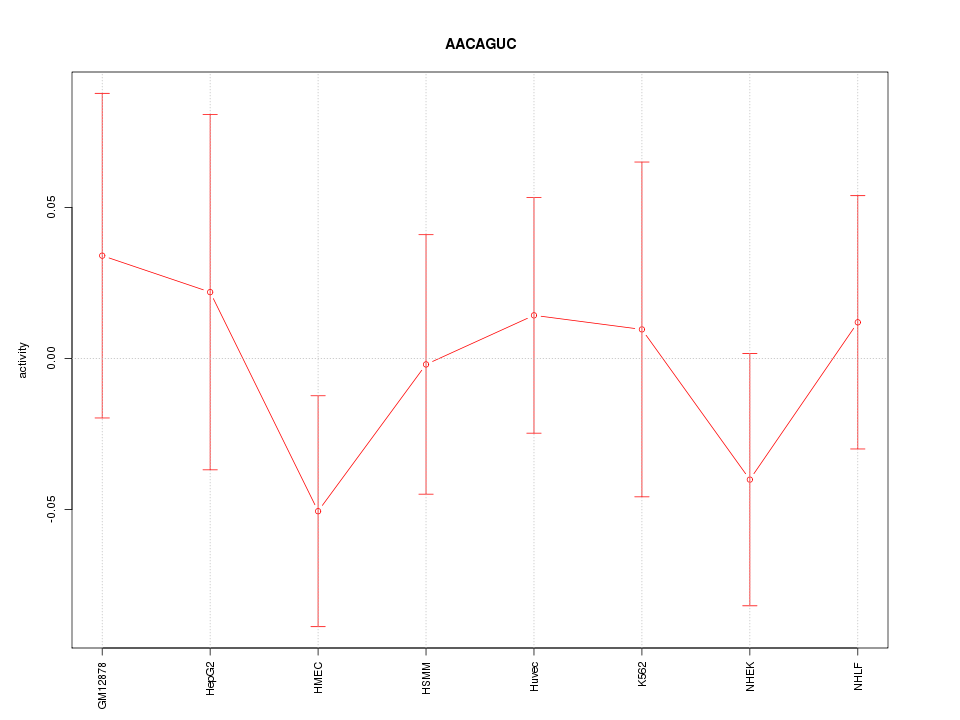
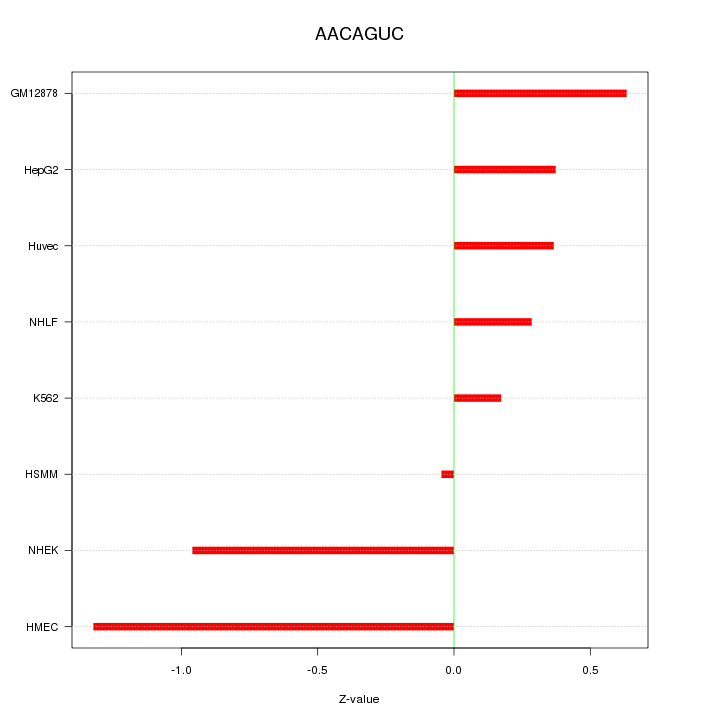
| 0.1 |
0.3 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 |
0.3 |
GO:0060292 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) long term synaptic depression(GO:0060292) |
| 0.1 |
0.3 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 |
0.2 |
GO:0070445 |
negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) regulation of photoreceptor cell differentiation(GO:0046532) male genitalia morphogenesis(GO:0048808) formation of anatomical boundary(GO:0048859) negative regulation of smooth muscle cell differentiation(GO:0051151) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) kidney mesenchymal cell proliferation(GO:0072135) metanephric mesenchymal cell proliferation involved in metanephros development(GO:0072136) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 |
0.2 |
GO:0001544 |
initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 |
0.1 |
GO:0032071 |
regulation of endodeoxyribonuclease activity(GO:0032071) negative regulation of helicase activity(GO:0051097) |
| 0.0 |
0.3 |
GO:0051387 |
negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 |
0.1 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.0 |
0.3 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.1 |
GO:0010586 |
miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 |
0.3 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.0 |
0.1 |
GO:0007109 |
obsolete cytokinesis, completion of separation(GO:0007109) protein hexamerization(GO:0034214) |
| 0.0 |
0.1 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.0 |
0.5 |
GO:0051319 |
mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 |
0.2 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.1 |
GO:1901724 |
response to heparin(GO:0071503) cellular response to heparin(GO:0071504) positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.0 |
0.1 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.1 |
GO:0060318 |
CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) definitive erythrocyte differentiation(GO:0060318) |
| 0.0 |
0.1 |
GO:0043045 |
DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 |
0.2 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.1 |
GO:0019852 |
L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 |
0.1 |
GO:0036037 |
CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.1 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.0 |
0.1 |
GO:0016265 |
obsolete death(GO:0016265) |
| 0.0 |
0.0 |
GO:0035120 |
post-embryonic appendage morphogenesis(GO:0035120) |
| 0.0 |
0.0 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 |
0.4 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.1 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.2 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |