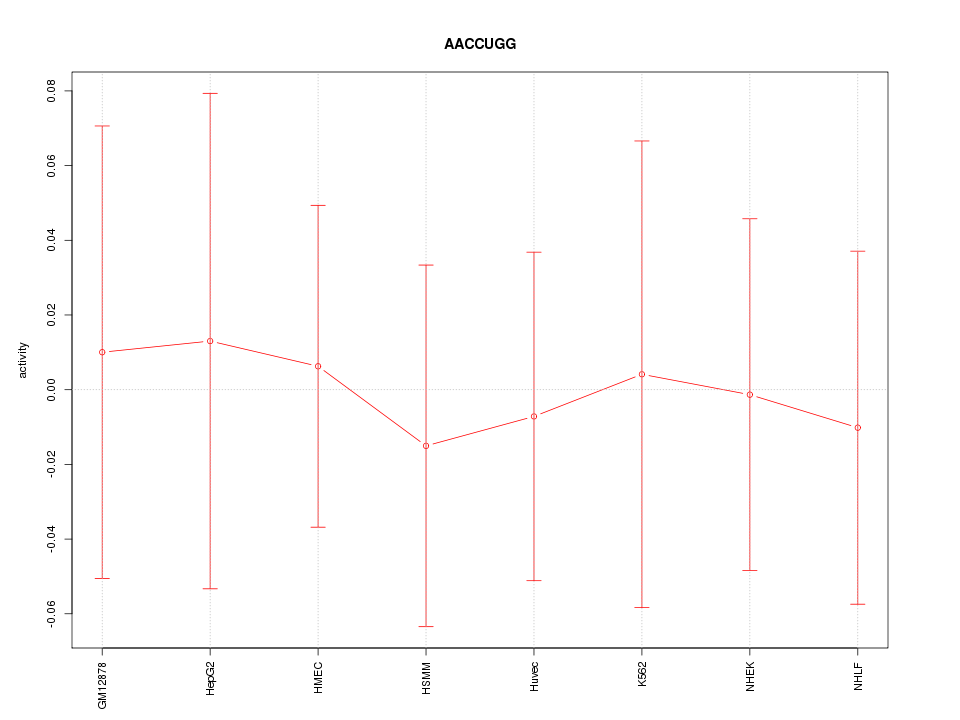
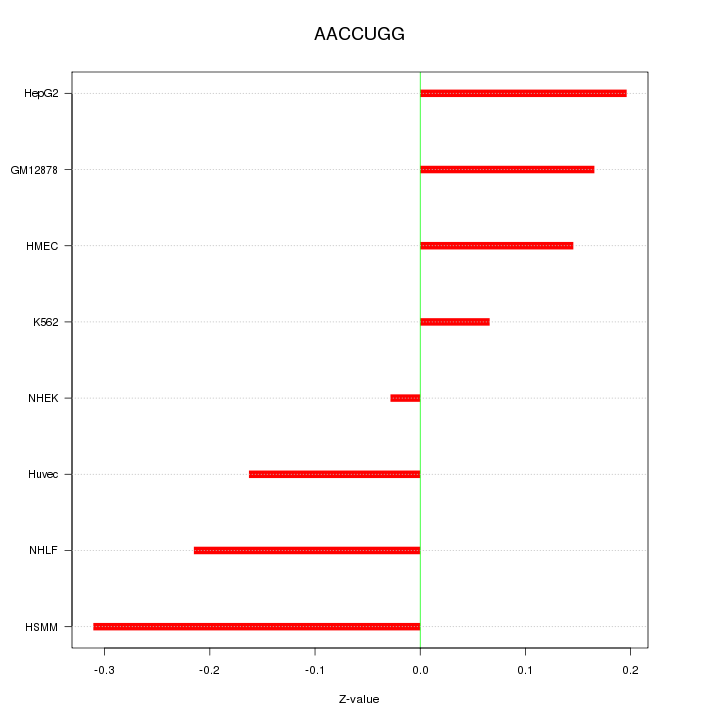
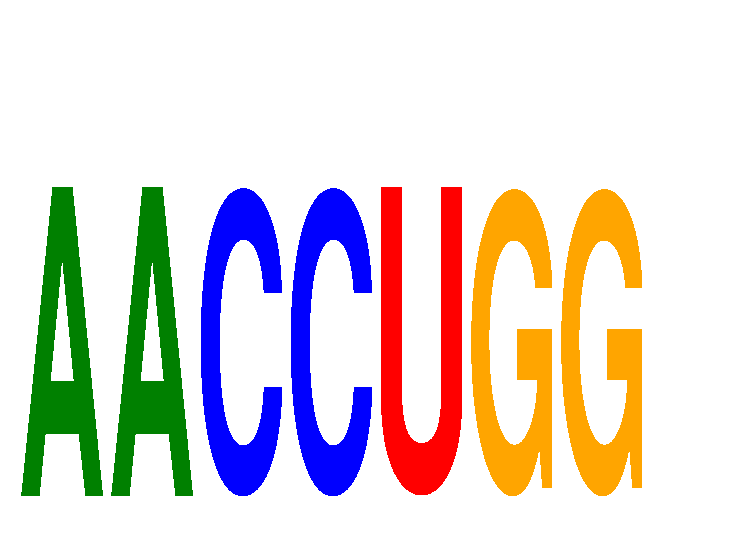|
chr11_-_115375107
|
0.092
|
ENST00000545380.1
ENST00000452722.3
ENST00000537058.1
ENST00000536727.1
ENST00000542447.2
ENST00000331581.6
|
CADM1
|
cell adhesion molecule 1
|
|
chr7_-_139876812
|
0.078
|
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A
|
|
chr18_+_55102917
|
0.064
|
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2
|
|
chr1_-_247094628
|
0.061
|
ENST00000366508.1
ENST00000326225.3
ENST00000391829.2
|
AHCTF1
|
AT hook containing transcription factor 1
|
|
chrX_+_123095155
|
0.054
|
ENST00000371160.1
ENST00000435103.1
|
STAG2
|
stromal antigen 2
|
|
chr19_+_34919257
|
0.051
|
ENST00000246548.4
ENST00000590048.2
|
UBA2
|
ubiquitin-like modifier activating enzyme 2
|
|
chr16_+_1203194
|
0.042
|
ENST00000348261.5
ENST00000358590.4
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr13_-_41240717
|
0.033
|
ENST00000379561.5
|
FOXO1
|
forkhead box O1
|
|
chr11_+_63706444
|
0.032
|
ENST00000377793.4
ENST00000456907.2
ENST00000539656.1
|
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit
|
|
chr17_-_48785216
|
0.030
|
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr19_+_4007644
|
0.028
|
ENST00000262971.2
|
PIAS4
|
protein inhibitor of activated STAT, 4
|
|
chr18_+_42260861
|
0.027
|
ENST00000282030.5
|
SETBP1
|
SET binding protein 1
|
|
chr3_-_189838670
|
0.026
|
ENST00000319332.5
|
LEPREL1
|
leprecan-like 1
|
|
chr9_-_38069208
|
0.021
|
ENST00000377707.3
ENST00000377700.4
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr11_-_129062093
|
0.017
|
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr8_-_103876965
|
0.016
|
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1
|
|
chr11_+_76571911
|
0.016
|
ENST00000534206.1
ENST00000532485.1
ENST00000526597.1
ENST00000533873.1
ENST00000538157.1
|
ACER3
|
alkaline ceramidase 3
|
|
chr12_-_123011536
|
0.015
|
ENST00000331738.7
ENST00000354654.2
|
RSRC2
|
arginine/serine-rich coiled-coil 2
|
|
chr4_+_184020398
|
0.014
|
ENST00000403733.3
ENST00000378925.3
|
WWC2
|
WW and C2 domain containing 2
|
|
chr12_+_67663056
|
0.012
|
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1
|
|
chr9_-_123964114
|
0.010
|
ENST00000373840.4
|
RAB14
|
RAB14, member RAS oncogene family
|
|
chr4_-_113437328
|
0.009
|
ENST00000313341.3
|
NEUROG2
|
neurogenin 2
|
|
chr15_-_27018175
|
0.008
|
ENST00000311550.5
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr14_+_57735614
|
0.008
|
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit
|
|
chr11_+_125462690
|
0.007
|
ENST00000392708.4
ENST00000529196.1
ENST00000531491.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic)
|
|
chr8_-_17104356
|
0.005
|
ENST00000361272.4
ENST00000523917.1
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr1_+_93811438
|
0.003
|
ENST00000370272.4
ENST00000370267.1
|
DR1
|
down-regulator of transcription 1, TBP-binding (negative cofactor 2)
|
|
chr2_+_210444142
|
0.002
|
ENST00000360351.4
ENST00000361559.4
|
MAP2
|
microtubule-associated protein 2
|
|
chr15_+_52121822
|
0.002
|
ENST00000558455.1
ENST00000308580.7
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|