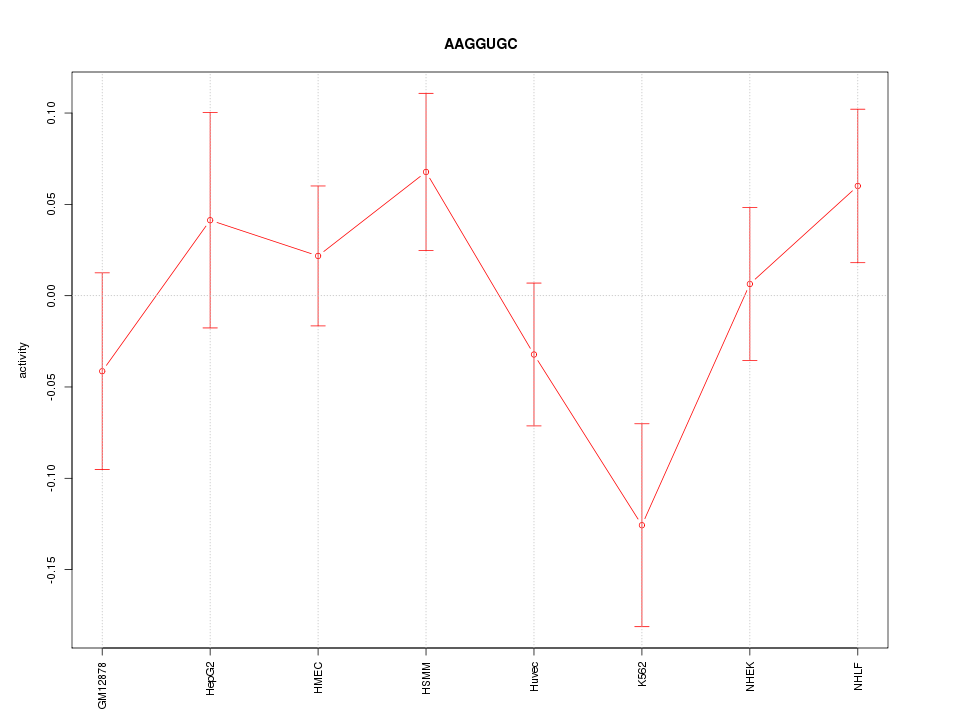
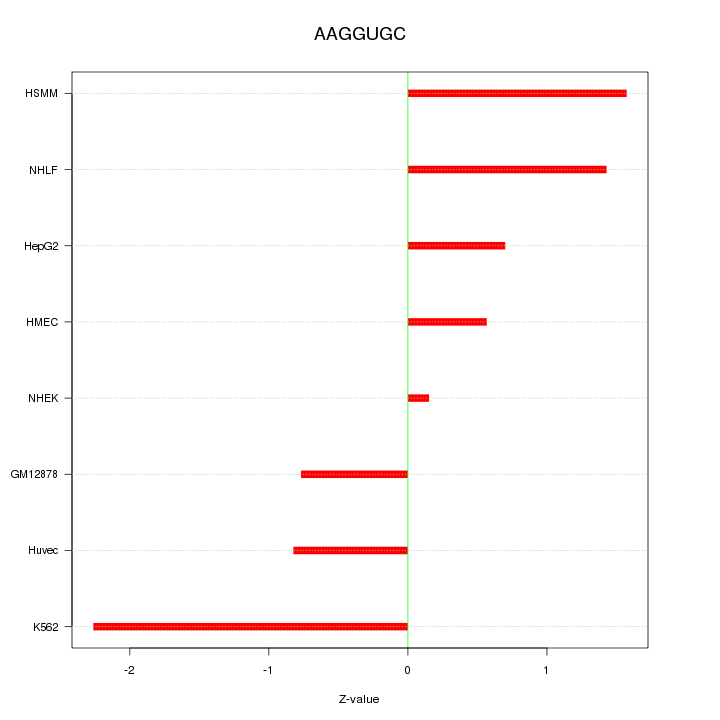
Motif ID: AAGGUGC
Z-value: 1.211
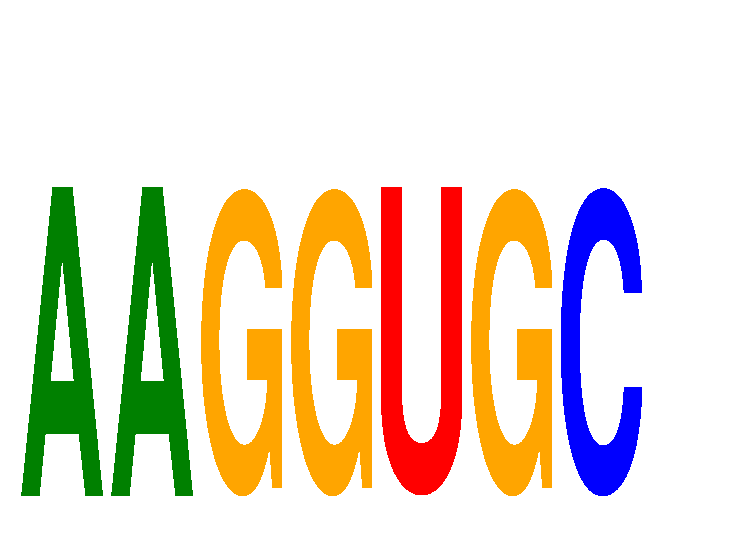
Mature miRNA associated with seed AAGGUGC:
| Name | miRBase Accession |
|---|---|
| hsa-miR-18a-5p | MIMAT0000072 |
| hsa-miR-18b-5p | MIMAT0001412 |
| hsa-miR-4735-3p | MIMAT0019861 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.5 | 3.2 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.6 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.2 | 0.8 | GO:0002331 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 2.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.8 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) |
| 0.1 | 0.3 | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.1 | 1.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 1.8 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 1.2 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.3 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.4 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 3.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 4.5 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.8 | 2.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.6 | 3.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 2.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.3 | 0.8 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.3 | 0.8 | GO:0050816 | beta-2 adrenergic receptor binding(GO:0031698) phosphothreonine binding(GO:0050816) |
| 0.2 | 1.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.4 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.6 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.1 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.8 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 2.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 1.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |