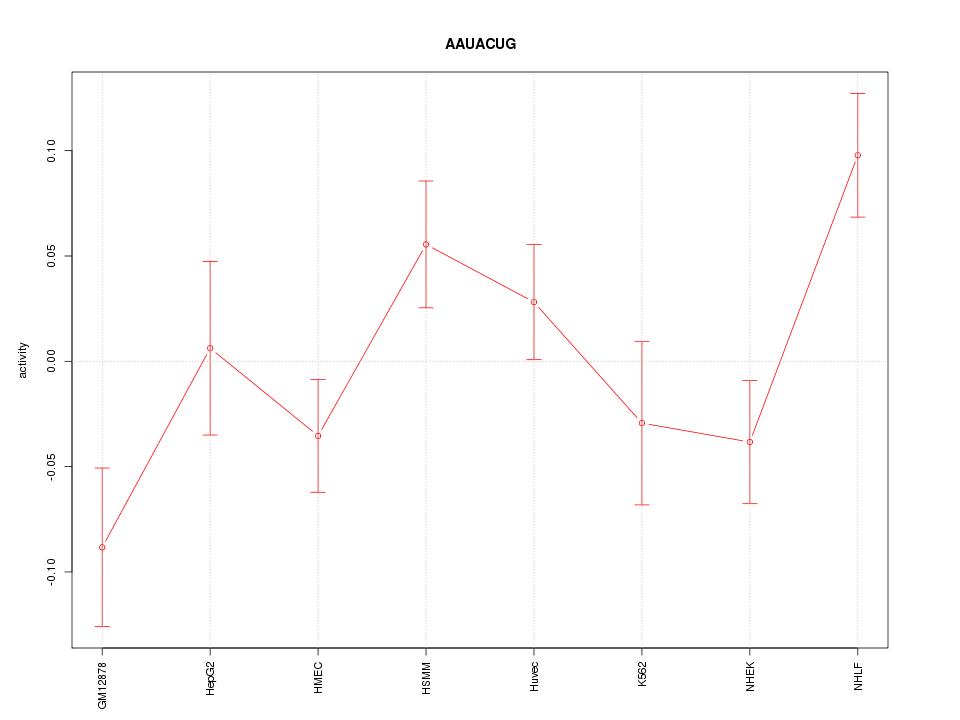
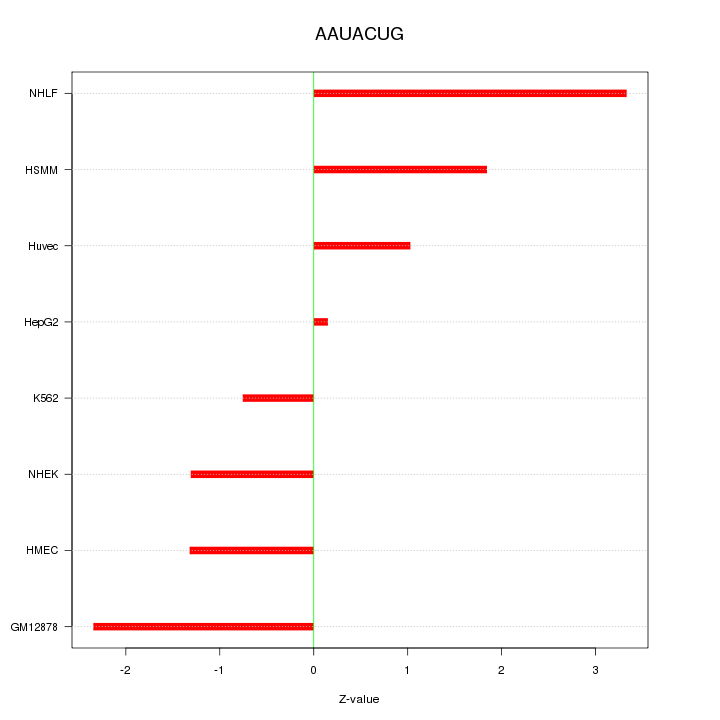
| 0.8 |
3.8 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.6 |
1.7 |
GO:0002887 |
negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) establishment of epithelial cell apical/basal polarity(GO:0045198) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.5 |
3.7 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.5 |
3.4 |
GO:0060414 |
aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.5 |
1.4 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.4 |
3.6 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.4 |
1.2 |
GO:0060574 |
trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 0.3 |
1.0 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.3 |
1.3 |
GO:0015825 |
L-serine transport(GO:0015825) |
| 0.3 |
1.0 |
GO:0021836 |
chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.3 |
1.8 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.3 |
0.9 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.3 |
2.1 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.2 |
0.2 |
GO:0060831 |
smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.2 |
1.0 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.2 |
1.6 |
GO:0022617 |
extracellular matrix disassembly(GO:0022617) |
| 0.2 |
1.8 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.2 |
0.5 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.2 |
0.5 |
GO:0007403 |
glial cell fate determination(GO:0007403) |
| 0.2 |
0.5 |
GO:0071918 |
urea transmembrane transport(GO:0071918) |
| 0.2 |
0.8 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.4 |
GO:0070092 |
regulation of glucagon secretion(GO:0070092) |
| 0.1 |
1.3 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.1 |
1.9 |
GO:0034405 |
response to fluid shear stress(GO:0034405) |
| 0.1 |
0.8 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 |
0.8 |
GO:0021800 |
cerebral cortex tangential migration(GO:0021800) |
| 0.1 |
1.2 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.1 |
1.9 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.1 |
1.0 |
GO:0051497 |
negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 |
2.4 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.1 |
0.4 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 |
0.6 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 |
0.2 |
GO:0051495 |
positive regulation of cytoskeleton organization(GO:0051495) |
| 0.1 |
0.7 |
GO:0042249 |
establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.1 |
0.2 |
GO:1903960 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.1 |
0.5 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 |
0.5 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.1 |
1.5 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.1 |
0.2 |
GO:0045578 |
progesterone secretion(GO:0042701) negative regulation of B cell differentiation(GO:0045578) regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 |
0.5 |
GO:0002063 |
chondrocyte development(GO:0002063) |
| 0.1 |
0.2 |
GO:0002124 |
territorial aggressive behavior(GO:0002124) brainstem development(GO:0003360) vocalization behavior(GO:0071625) |
| 0.1 |
0.5 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 |
0.3 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.1 |
0.3 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.1 |
0.3 |
GO:0046607 |
positive regulation of centrosome cycle(GO:0046607) |
| 0.1 |
0.6 |
GO:0021513 |
spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.1 |
0.6 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.1 |
0.1 |
GO:0008356 |
asymmetric cell division(GO:0008356) |
| 0.1 |
0.2 |
GO:0060174 |
limb bud formation(GO:0060174) |
| 0.1 |
1.3 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.1 |
0.2 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.1 |
0.3 |
GO:0032509 |
endosome transport via multivesicular body sorting pathway(GO:0032509) plasma membrane to endosome transport(GO:0048227) |
| 0.1 |
0.7 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.1 |
0.2 |
GO:0006679 |
glucosylceramide biosynthetic process(GO:0006679) |
| 0.1 |
0.2 |
GO:0030812 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.1 |
0.2 |
GO:0032707 |
negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 |
0.6 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 |
0.3 |
GO:0048146 |
fibroblast proliferation(GO:0048144) regulation of fibroblast proliferation(GO:0048145) positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 |
0.1 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.1 |
GO:0035408 |
histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 |
0.2 |
GO:0021873 |
forebrain neuroblast division(GO:0021873) neuronal stem cell division(GO:0036445) neuroblast division(GO:0055057) |
| 0.0 |
0.2 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 |
0.1 |
GO:0002378 |
immunoglobulin biosynthetic process(GO:0002378) protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.4 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
3.5 |
GO:0071772 |
BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 |
0.1 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.0 |
1.8 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
0.9 |
GO:0046847 |
filopodium assembly(GO:0046847) |
| 0.0 |
0.1 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.0 |
0.4 |
GO:0032429 |
regulation of phospholipase A2 activity(GO:0032429) |
| 0.0 |
0.2 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 |
0.4 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.7 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.0 |
0.5 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.0 |
0.1 |
GO:0001999 |
renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) |
| 0.0 |
0.3 |
GO:0008064 |
regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.0 |
0.3 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
0.0 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 |
2.4 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.7 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
0.2 |
GO:0002040 |
sprouting angiogenesis(GO:0002040) |
| 0.0 |
0.2 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.0 |
1.9 |
GO:0031032 |
actomyosin structure organization(GO:0031032) |
| 0.0 |
2.2 |
GO:0030814 |
regulation of cAMP metabolic process(GO:0030814) |
| 0.0 |
0.4 |
GO:0090174 |
organelle membrane fusion(GO:0090174) |
| 0.0 |
2.0 |
GO:0030041 |
actin filament polymerization(GO:0030041) |
| 0.0 |
0.3 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.7 |
GO:0001570 |
vasculogenesis(GO:0001570) |
| 0.0 |
0.0 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 |
0.6 |
GO:0001756 |
somitogenesis(GO:0001756) somite development(GO:0061053) |
| 0.0 |
0.2 |
GO:0043537 |
negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 |
0.8 |
GO:0051101 |
regulation of DNA binding(GO:0051101) |
| 0.0 |
0.6 |
GO:0014047 |
glutamate secretion(GO:0014047) |
| 0.0 |
0.6 |
GO:0006023 |
aminoglycan biosynthetic process(GO:0006023) |
| 0.0 |
0.2 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.0 |
0.1 |
GO:0019852 |
L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 |
3.4 |
GO:0030036 |
actin cytoskeleton organization(GO:0030036) |
| 0.0 |
0.1 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.0 |
0.8 |
GO:0034332 |
adherens junction organization(GO:0034332) |
| 0.0 |
0.2 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.6 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.0 |
0.3 |
GO:0050766 |
positive regulation of phagocytosis(GO:0050766) |
| 0.0 |
0.2 |
GO:0002675 |
positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 |
0.2 |
GO:0007176 |
regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 |
0.1 |
GO:0032000 |
positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 |
0.1 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
1.0 |
GO:0008277 |
regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 |
0.1 |
GO:0051205 |
protein insertion into membrane(GO:0051205) |
| 0.0 |
0.1 |
GO:0042517 |
positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 |
0.2 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |