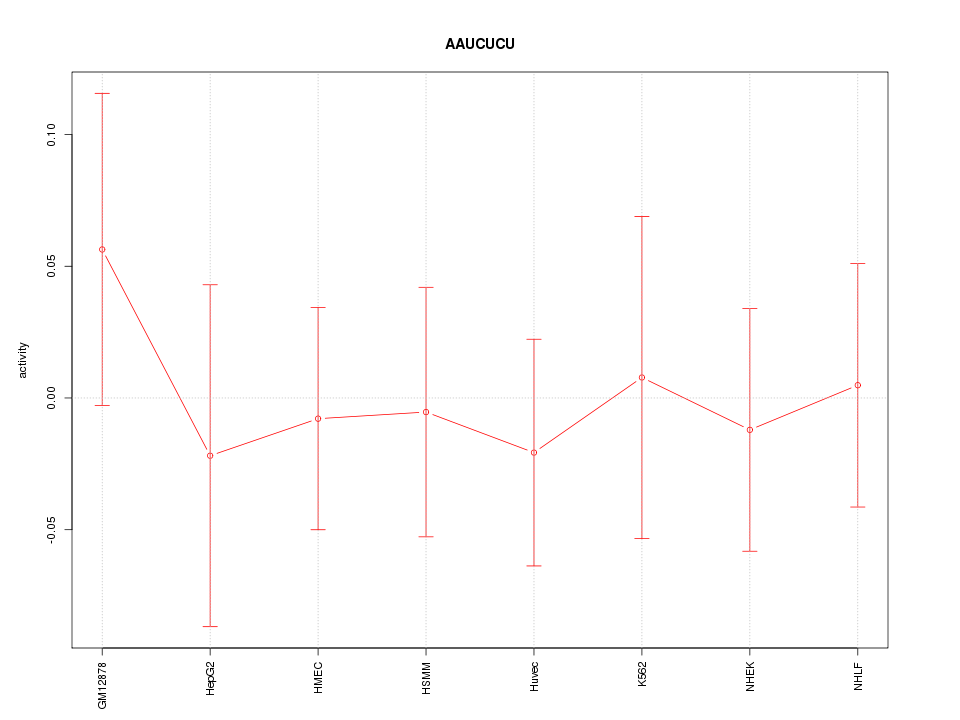
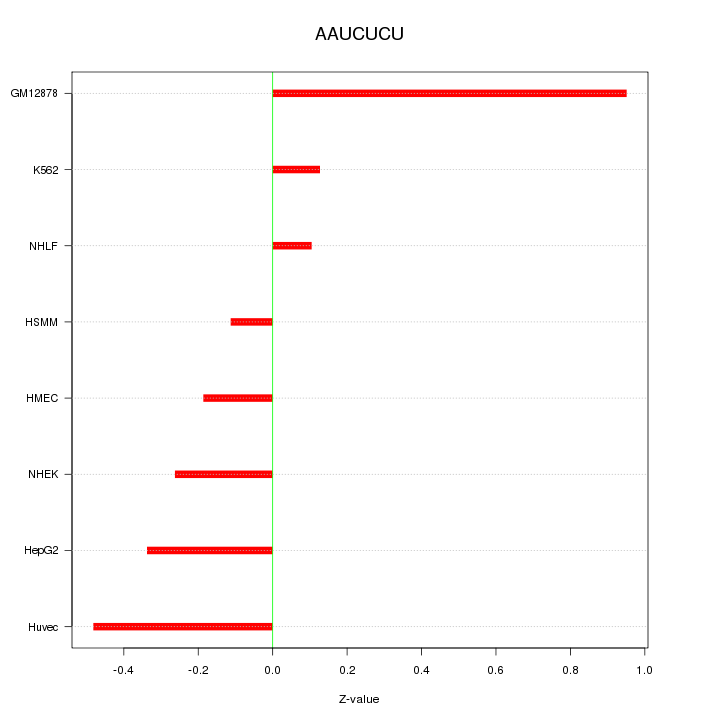
| 0.1 |
0.5 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 |
0.2 |
GO:0006848 |
pyruvate transport(GO:0006848) |
| 0.0 |
0.3 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.1 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 |
0.2 |
GO:0051096 |
meiotic mismatch repair(GO:0000710) positive regulation of helicase activity(GO:0051096) |
| 0.0 |
0.0 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.0 |
0.1 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.0 |
0.0 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.1 |
GO:0048892 |
sensory system development(GO:0048880) lateral line nerve development(GO:0048892) lateral line nerve glial cell differentiation(GO:0048895) lateral line system development(GO:0048925) lateral line nerve glial cell development(GO:0048937) iridophore differentiation(GO:0050935) |