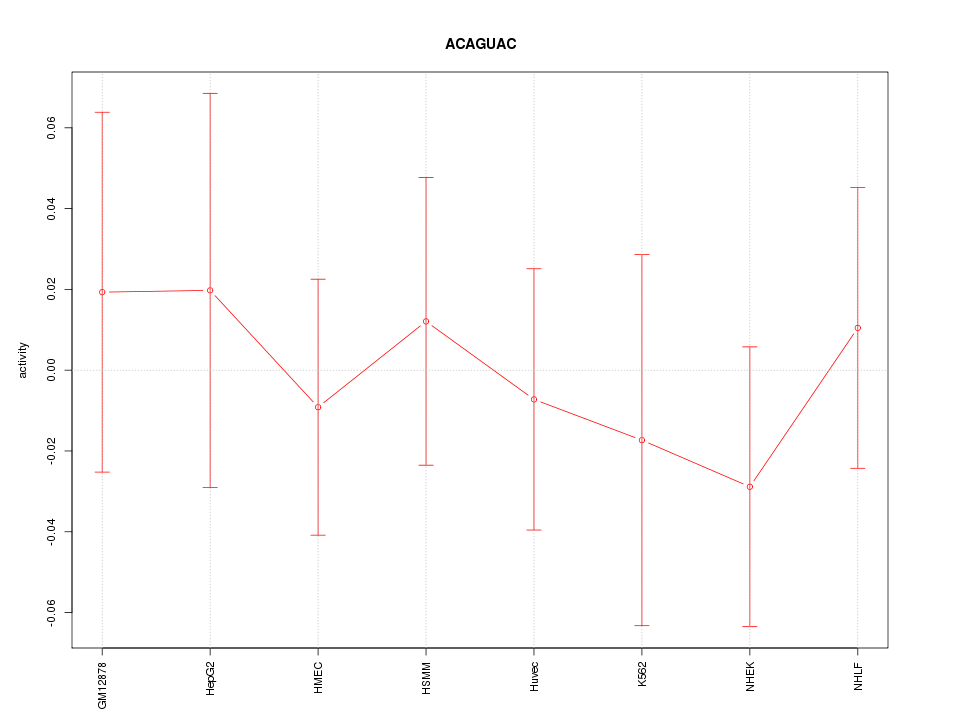
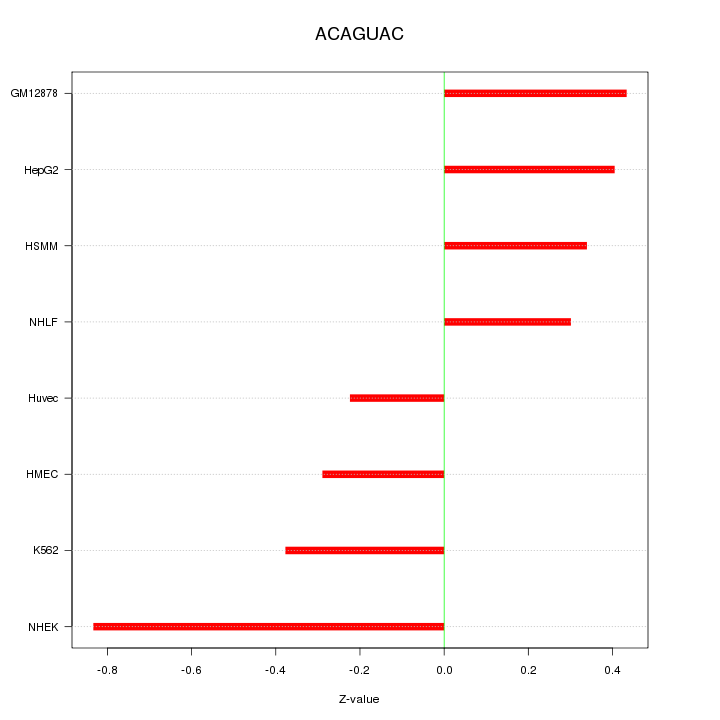
| 0.2 |
0.2 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.1 |
0.3 |
GO:0061298 |
progesterone secretion(GO:0042701) retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 |
0.4 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.1 |
0.5 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.2 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.1 |
0.3 |
GO:0060297 |
regulation of sarcomere organization(GO:0060297) |
| 0.1 |
0.2 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.1 |
0.2 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 |
0.2 |
GO:1904358 |
positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 |
0.1 |
GO:0070672 |
response to interleukin-12(GO:0070671) response to interleukin-15(GO:0070672) |
| 0.0 |
0.2 |
GO:0048669 |
collateral sprouting in absence of injury(GO:0048669) |
| 0.0 |
0.3 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 |
0.3 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 |
0.2 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.1 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.0 |
0.1 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 |
0.1 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.0 |
0.1 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 |
0.0 |
GO:0051573 |
negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 |
0.2 |
GO:0034616 |
response to laminar fluid shear stress(GO:0034616) |
| 0.0 |
0.1 |
GO:0006333 |
chromatin assembly or disassembly(GO:0006333) |
| 0.0 |
0.2 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 |
0.1 |
GO:0008218 |
bioluminescence(GO:0008218) |
| 0.0 |
0.1 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.1 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.1 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.1 |
GO:0048935 |
peripheral nervous system neuron development(GO:0048935) |
| 0.0 |
0.1 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
0.0 |
GO:0002887 |
negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) establishment of epithelial cell apical/basal polarity(GO:0045198) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 |
0.0 |
GO:0010834 |
obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 |
0.1 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.0 |
0.1 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.1 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.0 |
0.0 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.1 |
GO:0046967 |
cytosol to ER transport(GO:0046967) |
| 0.0 |
0.1 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 |
0.3 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.1 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 |
0.1 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.1 |
GO:0051571 |
positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 |
0.1 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 |
0.0 |
GO:0071635 |
negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 |
0.0 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.0 |
0.0 |
GO:0031396 |
regulation of protein ubiquitination(GO:0031396) |
| 0.0 |
0.3 |
GO:0046847 |
filopodium assembly(GO:0046847) |
| 0.0 |
0.1 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.0 |
GO:0035234 |
ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 |
0.0 |
GO:0043044 |
ATP-dependent chromatin remodeling(GO:0043044) |