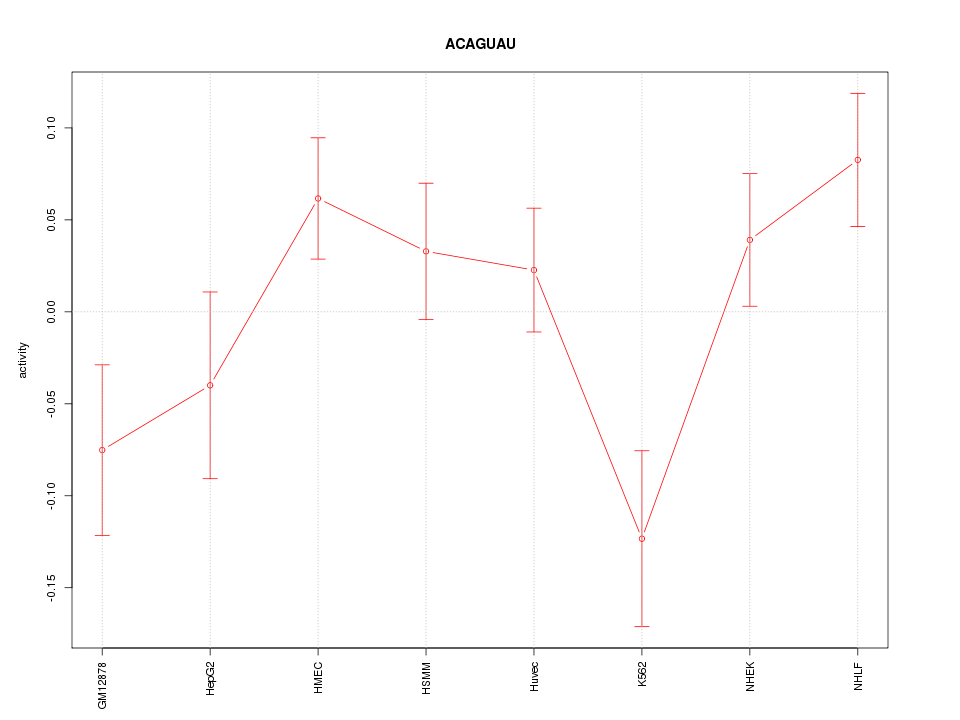
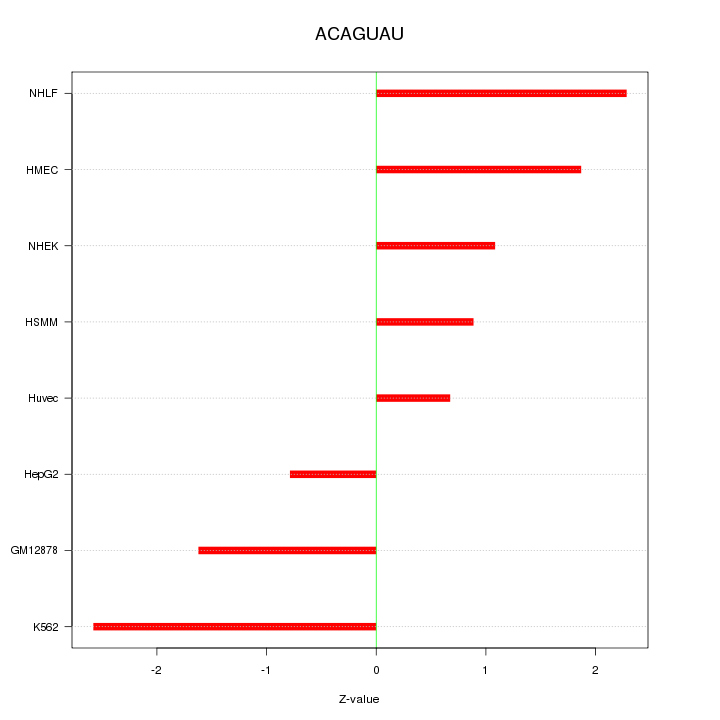
| 1.6 |
4.9 |
GO:0014029 |
neural crest formation(GO:0014029) neural crest cell fate commitment(GO:0014034) convergent extension involved in axis elongation(GO:0060028) cellular response to prostaglandin E stimulus(GO:0071380) response to heparin(GO:0071503) cellular response to heparin(GO:0071504) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 1.2 |
3.6 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 1.0 |
2.9 |
GO:0070092 |
regulation of glucagon secretion(GO:0070092) |
| 0.9 |
5.4 |
GO:0032926 |
negative regulation of gonadotropin secretion(GO:0032277) negative regulation of activin receptor signaling pathway(GO:0032926) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.8 |
2.3 |
GO:0002514 |
B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.6 |
2.3 |
GO:0051503 |
purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) |
| 0.6 |
1.7 |
GO:0008218 |
bioluminescence(GO:0008218) |
| 0.5 |
0.5 |
GO:0048369 |
lateral mesoderm development(GO:0048368) lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.5 |
4.1 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.4 |
2.8 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.4 |
1.8 |
GO:0048669 |
collateral sprouting in absence of injury(GO:0048669) |
| 0.3 |
0.9 |
GO:0043006 |
activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.3 |
1.2 |
GO:0031394 |
regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 |
1.2 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 |
1.1 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.2 |
0.7 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 |
3.6 |
GO:0060638 |
mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.2 |
0.4 |
GO:0002890 |
negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.2 |
1.0 |
GO:0044854 |
membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) regulation of dopamine receptor signaling pathway(GO:0060159) positive regulation of dopamine receptor signaling pathway(GO:0060161) caveola assembly(GO:0070836) membrane assembly(GO:0071709) |
| 0.2 |
0.5 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.2 |
0.5 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.1 |
1.2 |
GO:0044341 |
sodium-dependent phosphate transport(GO:0044341) |
| 0.1 |
0.4 |
GO:0045656 |
negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 |
1.5 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.1 |
0.4 |
GO:0032510 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 |
0.5 |
GO:0034959 |
neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 |
0.3 |
GO:0070672 |
response to interleukin-15(GO:0070672) |
| 0.1 |
0.6 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 |
0.3 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.1 |
1.5 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.1 |
0.4 |
GO:0048289 |
isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.1 |
0.6 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.1 |
0.3 |
GO:0071109 |
superior temporal gyrus development(GO:0071109) |
| 0.1 |
1.2 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.1 |
0.4 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.1 |
0.2 |
GO:0035234 |
ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 |
0.3 |
GO:0060649 |
nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.1 |
0.2 |
GO:1904358 |
positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 |
0.9 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.1 |
0.4 |
GO:0022038 |
corpus callosum development(GO:0022038) |
| 0.1 |
1.2 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.1 |
0.8 |
GO:0034405 |
response to fluid shear stress(GO:0034405) |
| 0.1 |
0.2 |
GO:0034204 |
lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 |
0.3 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
0.6 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.1 |
0.3 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.1 |
0.4 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.1 |
0.3 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.1 |
0.2 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 |
0.2 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 |
0.3 |
GO:0010766 |
negative regulation of sodium ion transport(GO:0010766) |
| 0.0 |
0.9 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
1.4 |
GO:0048286 |
lung alveolus development(GO:0048286) |
| 0.0 |
1.1 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.0 |
0.5 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.0 |
0.2 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.0 |
3.6 |
GO:0006939 |
smooth muscle contraction(GO:0006939) |
| 0.0 |
0.1 |
GO:0060297 |
regulation of sarcomere organization(GO:0060297) |
| 0.0 |
0.1 |
GO:0001999 |
renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) |
| 0.0 |
0.5 |
GO:0001953 |
negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 |
0.1 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 |
0.8 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.0 |
0.0 |
GO:0048875 |
surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 |
0.2 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
2.8 |
GO:0034332 |
adherens junction organization(GO:0034332) |
| 0.0 |
0.3 |
GO:0050910 |
detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 |
0.1 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 |
1.0 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
4.6 |
GO:0043281 |
regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043281) |
| 0.0 |
0.2 |
GO:0032472 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.3 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.9 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.2 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.2 |
GO:0014044 |
Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.7 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.1 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 |
0.2 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.4 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.0 |
0.1 |
GO:0018027 |
peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 |
0.1 |
GO:0033522 |
histone H2A ubiquitination(GO:0033522) |
| 0.0 |
0.1 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.0 |
0.6 |
GO:0002208 |
somatic recombination of immunoglobulin genes involved in immune response(GO:0002204) somatic diversification of immunoglobulins involved in immune response(GO:0002208) isotype switching(GO:0045190) |
| 0.0 |
0.4 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.0 |
0.6 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
1.0 |
GO:0007091 |
metaphase/anaphase transition of mitotic cell cycle(GO:0007091) regulation of mitotic sister chromatid separation(GO:0010965) regulation of mitotic sister chromatid segregation(GO:0033047) metaphase/anaphase transition of cell cycle(GO:0044784) mitotic sister chromatid separation(GO:0051306) |
| 0.0 |
0.0 |
GO:0002829 |
negative regulation of type 2 immune response(GO:0002829) |
| 0.0 |
1.6 |
GO:0030308 |
negative regulation of cell growth(GO:0030308) |
| 0.0 |
0.1 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 |
0.2 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.8 |
GO:0007623 |
circadian rhythm(GO:0007623) |
| 0.0 |
0.0 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 |
0.1 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 |
0.3 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.0 |
0.2 |
GO:0006305 |
DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 |
0.6 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.2 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |