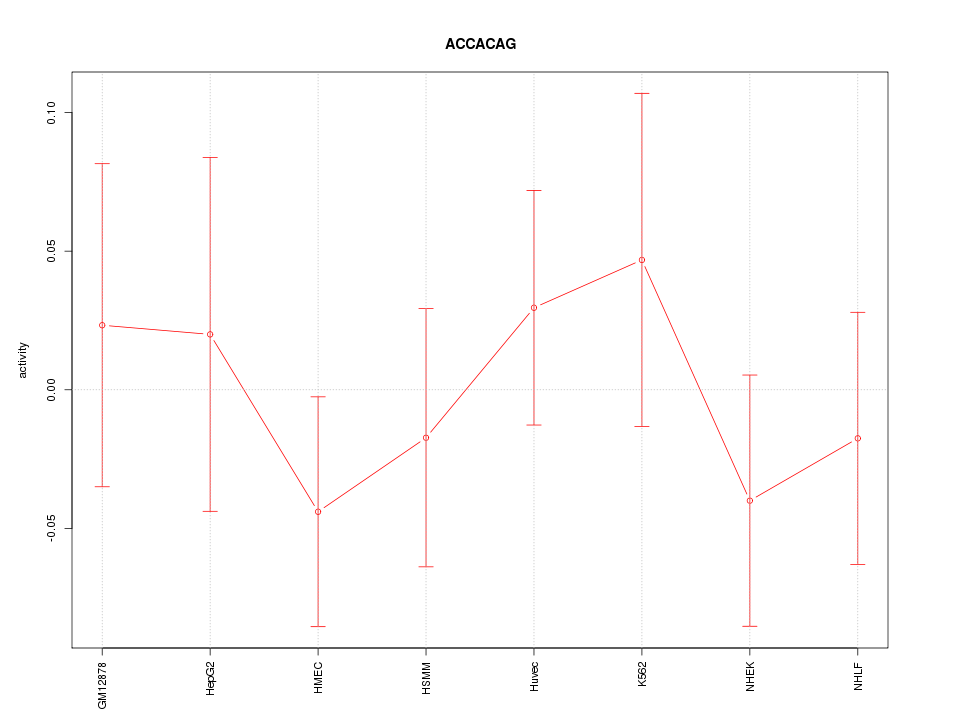
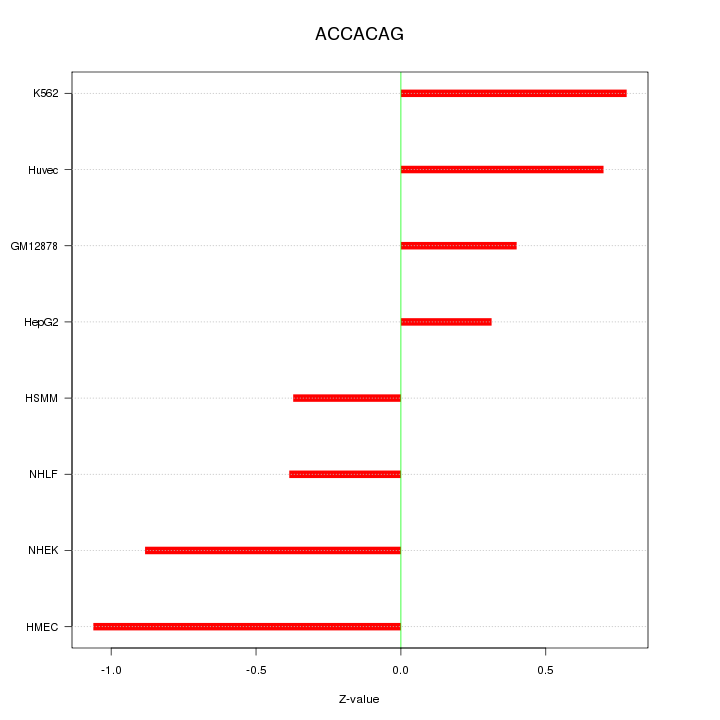
| 0.1 |
0.2 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 |
0.1 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.4 |
GO:0043306 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.1 |
GO:0055005 |
optic placode formation involved in camera-type eye formation(GO:0046619) ventricular cardiac myofibril assembly(GO:0055005) regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 |
0.2 |
GO:0048935 |
peripheral nervous system neuron development(GO:0048935) |
| 0.0 |
0.2 |
GO:0060292 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) long term synaptic depression(GO:0060292) |
| 0.0 |
0.1 |
GO:0046671 |
regulation of nitrogen utilization(GO:0006808) positive regulation of neuron maturation(GO:0014042) nitrogen utilization(GO:0019740) cochlear nucleus development(GO:0021747) negative regulation of cellular pH reduction(GO:0032848) glial cell apoptotic process(GO:0034349) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) positive regulation of cell maturation(GO:1903431) |
| 0.0 |
0.1 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.0 |
0.1 |
GO:1901142 |
insulin processing(GO:0030070) insulin metabolic process(GO:1901142) |
| 0.0 |
0.1 |
GO:0032071 |
regulation of endodeoxyribonuclease activity(GO:0032071) negative regulation of helicase activity(GO:0051097) |
| 0.0 |
0.1 |
GO:0006542 |
glutamine biosynthetic process(GO:0006542) |
| 0.0 |
0.1 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.3 |
GO:0043584 |
nose development(GO:0043584) |
| 0.0 |
0.1 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.1 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.1 |
GO:0032916 |
transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 |
0.1 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.0 |
0.2 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 |
0.1 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 |
0.2 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.0 |
GO:0007518 |
myoblast fate determination(GO:0007518) |
| 0.0 |
0.1 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.2 |
GO:0033261 |
obsolete regulation of S phase(GO:0033261) |
| 0.0 |
0.1 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 |
0.1 |
GO:0060676 |
ureteric bud formation(GO:0060676) mesonephric tubule formation(GO:0072172) |
| 0.0 |
0.1 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 |
0.0 |
GO:0001692 |
histamine metabolic process(GO:0001692) |
| 0.0 |
0.1 |
GO:0032876 |
regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |