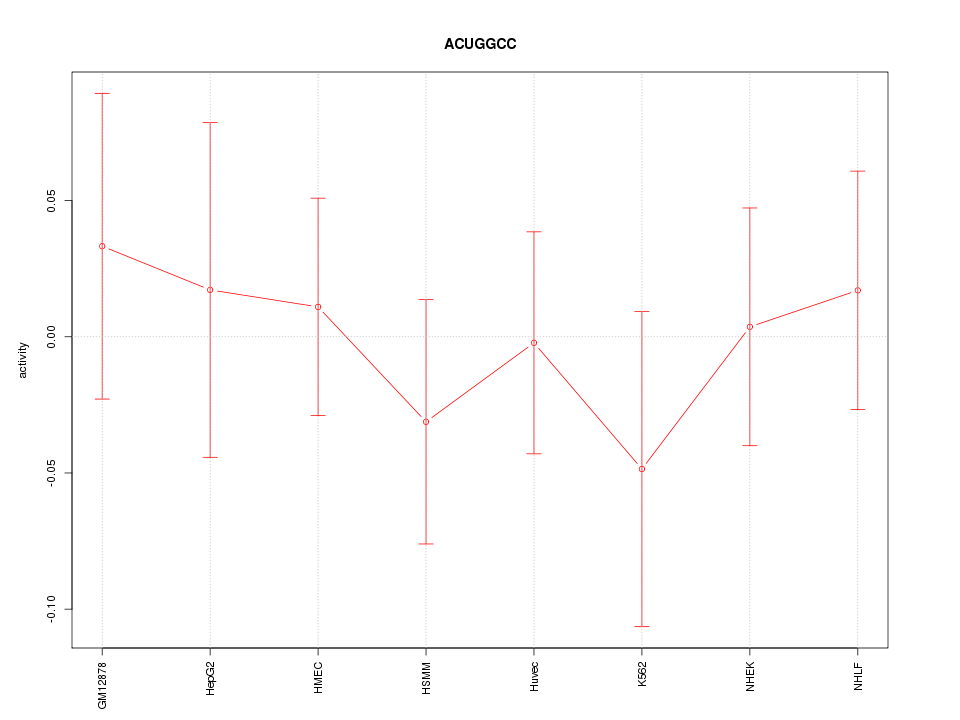
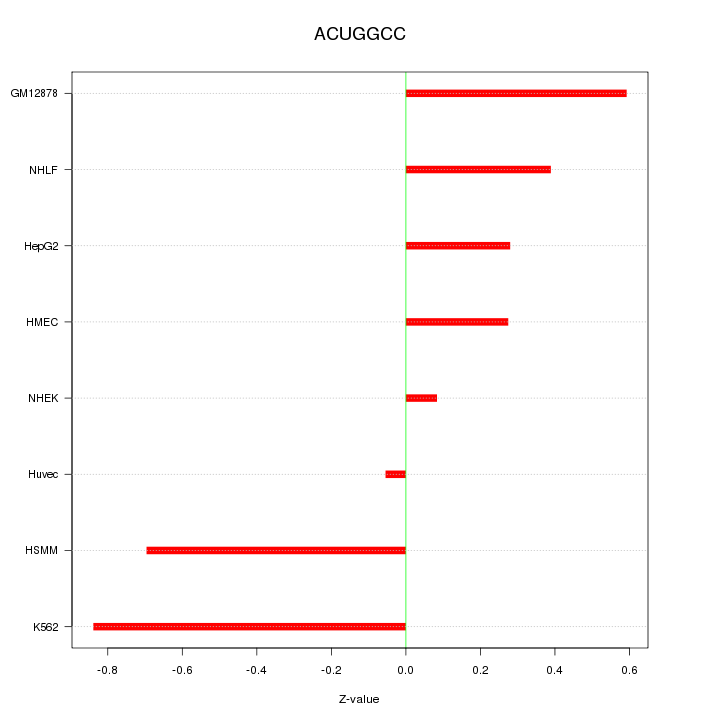
|
chr1_+_183605200
|
0.532
|
ENST00000304685.4
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr1_+_182992545
|
0.383
|
ENST00000258341.4
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr3_-_124774802
|
0.376
|
ENST00000311127.4
|
HEG1
|
heart development protein with EGF-like domains 1
|
|
chr6_-_29527702
|
0.362
|
ENST00000377050.4
|
UBD
|
ubiquitin D
|
|
chr11_-_115375107
|
0.357
|
ENST00000545380.1
ENST00000452722.3
ENST00000537058.1
ENST00000536727.1
ENST00000542447.2
ENST00000331581.6
|
CADM1
|
cell adhesion molecule 1
|
|
chr15_-_61521495
|
0.335
|
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A
|
|
chr12_+_94542459
|
0.332
|
ENST00000258526.4
|
PLXNC1
|
plexin C1
|
|
chr7_-_26904317
|
0.294
|
ENST00000345317.2
|
SKAP2
|
src kinase associated phosphoprotein 2
|
|
chr13_+_98794810
|
0.292
|
ENST00000595437.1
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr11_+_128563652
|
0.291
|
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor
|
|
chr10_+_97515409
|
0.235
|
ENST00000371207.3
ENST00000543964.1
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1
|
|
chr22_-_39548627
|
0.234
|
ENST00000216133.5
|
CBX7
|
chromobox homolog 7
|
|
chr20_-_14318248
|
0.229
|
ENST00000378053.3
ENST00000341420.4
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr6_-_16761678
|
0.220
|
ENST00000244769.4
ENST00000436367.1
|
ATXN1
|
ataxin 1
|
|
chr3_-_123603137
|
0.211
|
ENST00000360304.3
ENST00000359169.1
ENST00000346322.5
ENST00000360772.3
|
MYLK
|
myosin light chain kinase
|
|
chr4_-_23891693
|
0.181
|
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr1_-_92351769
|
0.176
|
ENST00000212355.4
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr3_-_57199397
|
0.174
|
ENST00000296318.7
|
IL17RD
|
interleukin 17 receptor D
|
|
chr2_-_208030647
|
0.172
|
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr16_-_4166186
|
0.161
|
ENST00000294016.3
|
ADCY9
|
adenylate cyclase 9
|
|
chr6_+_86159821
|
0.145
|
ENST00000369651.3
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr10_+_70480963
|
0.143
|
ENST00000265872.6
ENST00000535016.1
ENST00000538031.1
ENST00000543719.1
ENST00000539539.1
ENST00000543225.1
ENST00000536012.1
ENST00000494903.2
|
CCAR1
|
cell division cycle and apoptosis regulator 1
|
|
chr10_+_89622870
|
0.138
|
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog
|
|
chr6_-_75994536
|
0.137
|
ENST00000475111.2
ENST00000230461.6
|
TMEM30A
|
transmembrane protein 30A
|
|
chr8_-_57123815
|
0.136
|
ENST00000316981.3
ENST00000423799.2
ENST00000429357.2
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr17_-_38020392
|
0.133
|
ENST00000346872.3
ENST00000439167.2
ENST00000377945.3
ENST00000394189.2
ENST00000377944.3
ENST00000377958.2
ENST00000535189.1
ENST00000377952.2
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos)
|
|
chr20_-_4982132
|
0.126
|
ENST00000338244.1
ENST00000424750.2
|
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2
|
|
chr14_+_57046500
|
0.121
|
ENST00000261556.6
|
TMEM260
|
transmembrane protein 260
|
|
chr17_-_66287257
|
0.120
|
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6
|
|
chr18_+_55711575
|
0.112
|
ENST00000356462.6
ENST00000400345.3
ENST00000589054.1
ENST00000256832.7
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase
|
|
chr1_+_183155373
|
0.106
|
ENST00000493293.1
ENST00000264144.4
|
LAMC2
|
laminin, gamma 2
|
|
chr3_+_132136331
|
0.105
|
ENST00000260818.6
|
DNAJC13
|
DnaJ (Hsp40) homolog, subfamily C, member 13
|
|
chr2_+_204192942
|
0.099
|
ENST00000295851.5
ENST00000261017.5
|
ABI2
|
abl-interactor 2
|
|
chr6_-_47277634
|
0.098
|
ENST00000296861.2
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr12_+_11802753
|
0.098
|
ENST00000396373.4
|
ETV6
|
ets variant 6
|
|
chr11_-_27494279
|
0.091
|
ENST00000379214.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr12_-_57030115
|
0.087
|
ENST00000379441.3
ENST00000179765.5
ENST00000551812.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr8_-_93115445
|
0.084
|
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_+_38846101
|
0.078
|
ENST00000274276.3
|
OSMR
|
oncostatin M receptor
|
|
chr12_-_31744031
|
0.076
|
ENST00000389082.5
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr5_+_133861790
|
0.074
|
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2
|
|
chr22_+_24666763
|
0.069
|
ENST00000437398.1
ENST00000421374.1
ENST00000314328.9
ENST00000541492.1
|
SPECC1L
|
sperm antigen with calponin homology and coiled-coil domains 1-like
|
|
chr2_-_161350305
|
0.068
|
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr13_+_111365602
|
0.068
|
ENST00000333219.7
|
ING1
|
inhibitor of growth family, member 1
|
|
chr11_-_46940074
|
0.059
|
ENST00000378623.1
ENST00000534404.1
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr1_+_77333117
|
0.057
|
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr20_+_57226284
|
0.057
|
ENST00000458280.1
ENST00000355957.5
ENST00000361770.5
ENST00000312283.8
ENST00000412911.1
ENST00000359617.4
ENST00000371141.4
|
STX16
|
syntaxin 16
|
|
chrX_+_110339439
|
0.053
|
ENST00000372010.1
ENST00000519681.1
ENST00000372007.5
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr4_-_105416039
|
0.053
|
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4
|
|
chr1_+_151584544
|
0.050
|
ENST00000458013.2
ENST00000368843.3
|
SNX27
|
sorting nexin family member 27
|
|
chr1_-_169863016
|
0.049
|
ENST00000367772.4
ENST00000367771.6
|
SCYL3
|
SCY1-like 3 (S. cerevisiae)
|
|
chr12_-_111021110
|
0.044
|
ENST00000354300.3
|
PPTC7
|
PTC7 protein phosphatase homolog (S. cerevisiae)
|
|
chr18_+_46065393
|
0.042
|
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr1_+_203764742
|
0.040
|
ENST00000432282.1
ENST00000453771.1
ENST00000367214.1
ENST00000367212.3
ENST00000332127.4
|
ZC3H11A
|
zinc finger CCCH-type containing 11A
|
|
chr14_+_23775971
|
0.039
|
ENST00000250405.5
|
BCL2L2
|
BCL2-like 2
|
|
chr11_+_69455855
|
0.038
|
ENST00000227507.2
ENST00000536559.1
|
CCND1
|
cyclin D1
|
|
chr17_+_61699766
|
0.033
|
ENST00000579585.1
ENST00000584573.1
ENST00000361733.3
ENST00000361357.3
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr19_+_797392
|
0.032
|
ENST00000350092.4
ENST00000349038.4
ENST00000586481.1
ENST00000585535.1
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr15_+_64443905
|
0.031
|
ENST00000325881.4
|
SNX22
|
sorting nexin 22
|
|
chr1_-_67519782
|
0.028
|
ENST00000235345.5
|
SLC35D1
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1
|
|
chr14_-_68283291
|
0.028
|
ENST00000555452.1
ENST00000347230.4
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr11_-_77532050
|
0.028
|
ENST00000308488.6
|
RSF1
|
remodeling and spacing factor 1
|
|
chr22_+_40390930
|
0.027
|
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr5_+_172483347
|
0.027
|
ENST00000522692.1
ENST00000296953.2
ENST00000540014.1
ENST00000520420.1
|
CREBRF
|
CREB3 regulatory factor
|
|
chrX_-_47479246
|
0.027
|
ENST00000295987.7
ENST00000340666.4
|
SYN1
|
synapsin I
|
|
chr10_-_99052382
|
0.026
|
ENST00000466484.1
ENST00000358531.4
ENST00000453547.2
ENST00000316676.8
ENST00000358308.3
|
ARHGAP19
ARHGAP19-SLIT1
|
Rho GTPase activating protein 19
ARHGAP19-SLIT1 readthrough (NMD candidate)
|
|
chr3_-_52090461
|
0.023
|
ENST00000296483.6
ENST00000495880.1
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr2_+_113931513
|
0.021
|
ENST00000245796.6
ENST00000441564.3
|
PSD4
|
pleckstrin and Sec7 domain containing 4
|
|
chr2_+_29204161
|
0.018
|
ENST00000379558.4
ENST00000403861.2
|
FAM179A
|
family with sequence similarity 179, member A
|
|
chr9_-_135819987
|
0.018
|
ENST00000298552.3
ENST00000403810.1
|
TSC1
|
tuberous sclerosis 1
|
|
chr20_+_42086525
|
0.017
|
ENST00000244020.3
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chrX_-_40594755
|
0.016
|
ENST00000324817.1
|
MED14
|
mediator complex subunit 14
|
|
chr10_-_38146510
|
0.015
|
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248
|
|
chr11_-_124670550
|
0.014
|
ENST00000239614.4
|
MSANTD2
|
Myb/SANT-like DNA-binding domain containing 2
|
|
chr2_-_69870835
|
0.014
|
ENST00000409085.4
ENST00000406297.3
|
AAK1
|
AP2 associated kinase 1
|
|
chr11_+_118307179
|
0.014
|
ENST00000534358.1
ENST00000531904.2
ENST00000389506.5
ENST00000354520.4
|
KMT2A
|
lysine (K)-specific methyltransferase 2A
|
|
chr9_+_115983808
|
0.014
|
ENST00000374210.6
ENST00000374212.4
|
SLC31A1
|
solute carrier family 31 (copper transporter), member 1
|
|
chr3_+_71803201
|
0.014
|
ENST00000304411.2
|
GPR27
|
G protein-coupled receptor 27
|
|
chr2_-_131850951
|
0.013
|
ENST00000409185.1
ENST00000389915.3
|
FAM168B
|
family with sequence similarity 168, member B
|
|
chr1_-_150552006
|
0.013
|
ENST00000307940.3
ENST00000369026.2
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr4_-_87281224
|
0.010
|
ENST00000395169.3
ENST00000395161.2
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr3_-_128902729
|
0.009
|
ENST00000451728.2
ENST00000446936.2
ENST00000502976.1
ENST00000500450.2
ENST00000441626.2
|
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein
|
|
chr17_+_61627814
|
0.008
|
ENST00000310827.4
ENST00000431926.1
ENST00000415273.2
|
DCAF7
|
DDB1 and CUL4 associated factor 7
|
|
chr7_-_143059845
|
0.005
|
ENST00000443739.2
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr2_+_48541776
|
0.005
|
ENST00000413569.1
ENST00000340553.3
|
FOXN2
|
forkhead box N2
|
|
chr11_+_33563821
|
0.005
|
ENST00000321505.4
ENST00000265654.5
ENST00000389726.3
|
KIAA1549L
|
KIAA1549-like
|
|
chr12_-_42538657
|
0.005
|
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr17_+_48796905
|
0.004
|
ENST00000505658.1
ENST00000393227.2
ENST00000240304.1
ENST00000311571.3
ENST00000505619.1
ENST00000544170.1
ENST00000510984.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr14_-_27066636
|
0.003
|
ENST00000267422.7
ENST00000344429.5
ENST00000574031.1
ENST00000465357.2
ENST00000547619.1
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr3_-_52312636
|
0.003
|
ENST00000296490.3
|
WDR82
|
WD repeat domain 82
|
|
chr16_+_56225248
|
0.002
|
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr22_-_32146106
|
0.001
|
ENST00000327423.6
ENST00000397493.2
ENST00000434485.1
ENST00000412743.1
|
PRR14L
|
proline rich 14-like
|
|
chr17_+_8213590
|
0.000
|
ENST00000361926.3
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr19_-_14629224
|
0.000
|
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|