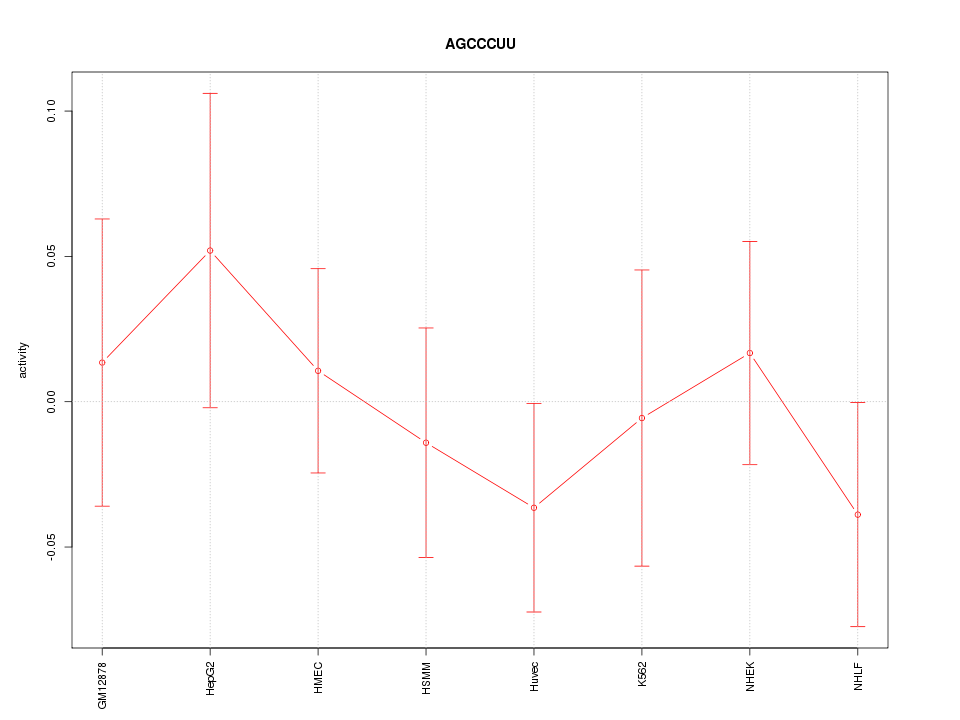
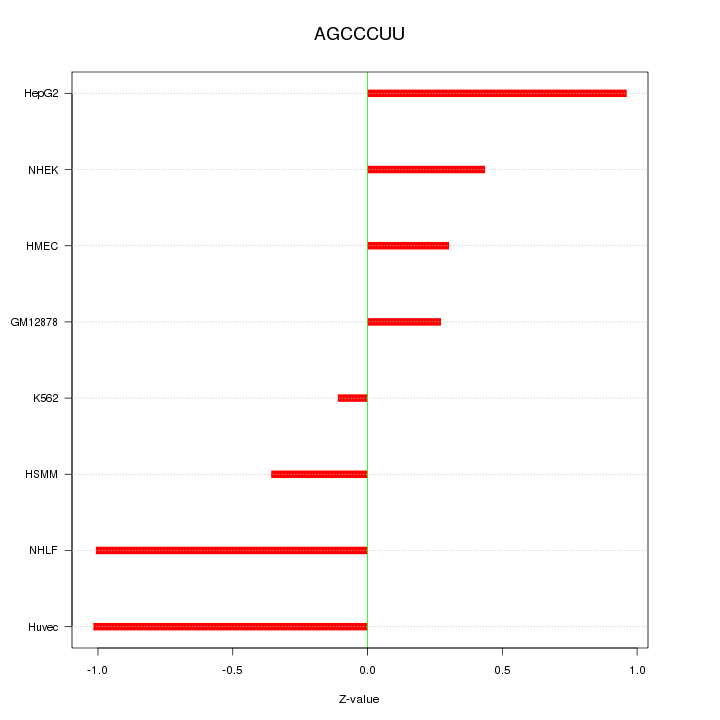
Motif ID: AGCCCUU
Z-value: 0.659

Mature miRNA associated with seed AGCCCUU:
| Name | miRBase Accession |
|---|---|
| hsa-miR-129-1-3p | MIMAT0004548 |
| hsa-miR-129-2-3p | MIMAT0004605 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.3 | GO:2000051 | pericardium morphogenesis(GO:0003344) post-embryonic appendage morphogenesis(GO:0035120) trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) mammary placode formation(GO:0060596) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.5 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.3 | GO:0009642 | response to light intensity(GO:0009642) response to vitamin E(GO:0033197) |
| 0.1 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.2 | GO:0071883 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.9 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 1.6 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 1.6 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.1 | 1.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.0 | 0.4 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.2 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.2 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase inhibitor activity(GO:0008073) ornithine decarboxylase activator activity(GO:0042978) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |