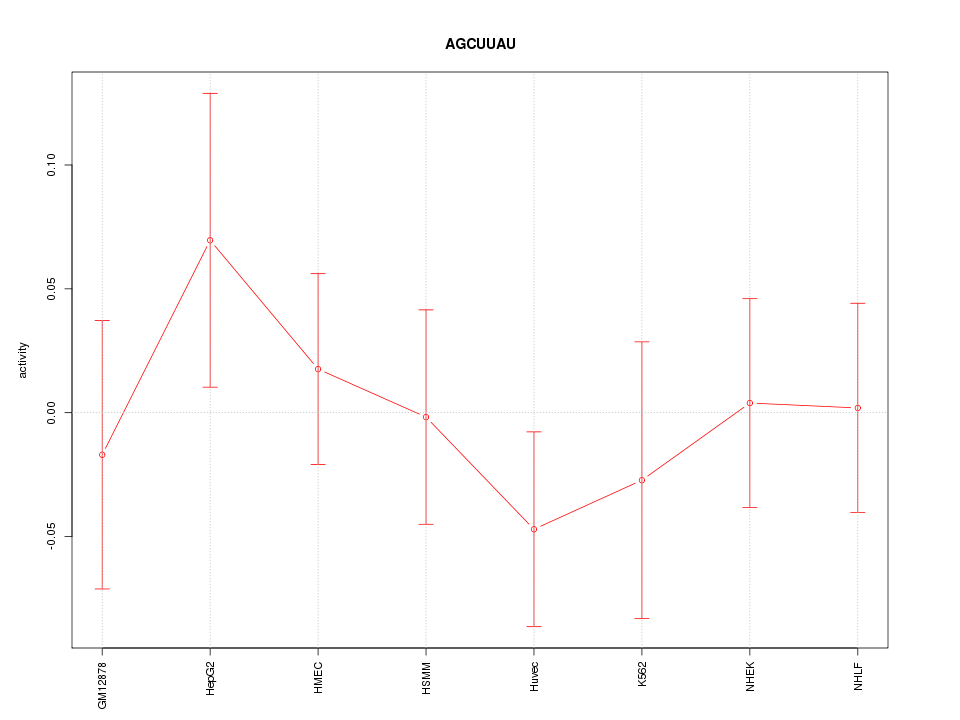
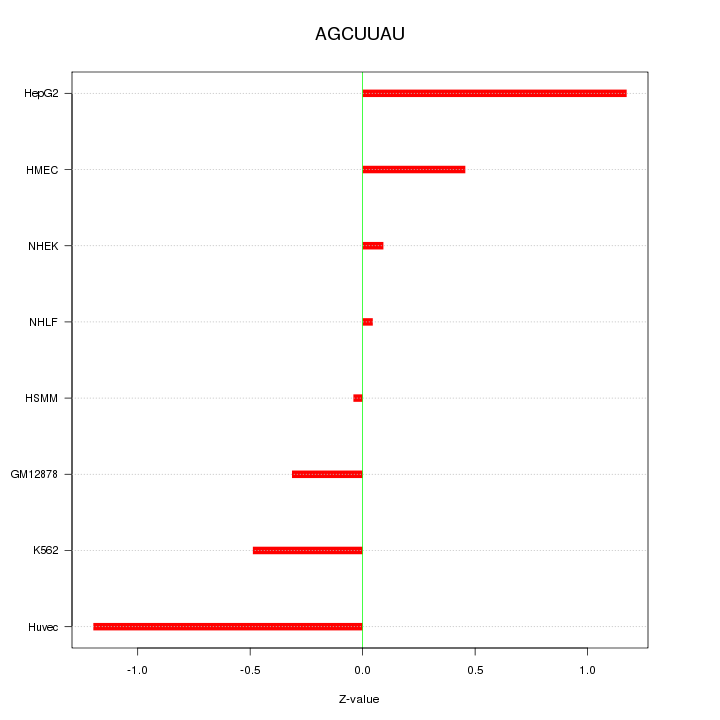
Motif ID: AGCUUAU
Z-value: 0.649

Mature miRNA associated with seed AGCUUAU:
| Name | miRBase Accession |
|---|---|
| hsa-miR-21-5p | MIMAT0000076 |
| hsa-miR-590-5p | MIMAT0003258 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 0.9 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 0.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.8 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 0.7 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.2 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.2 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.4 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.2 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.4 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.0 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.1 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.2 | GO:0060123 | female genitalia development(GO:0030540) regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 2.0 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.2 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 2.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |