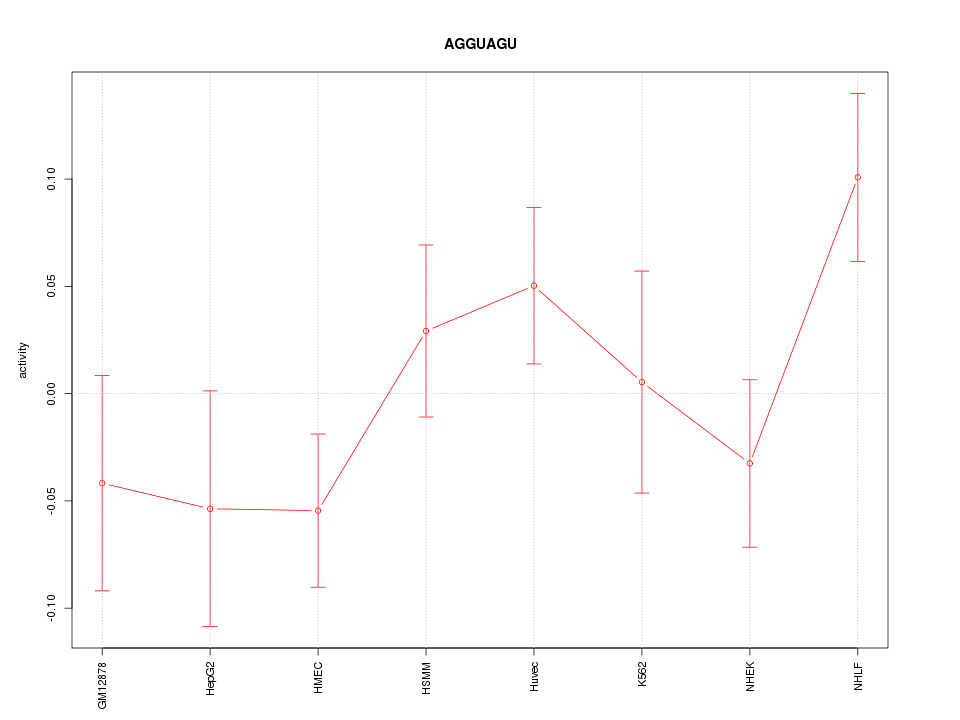
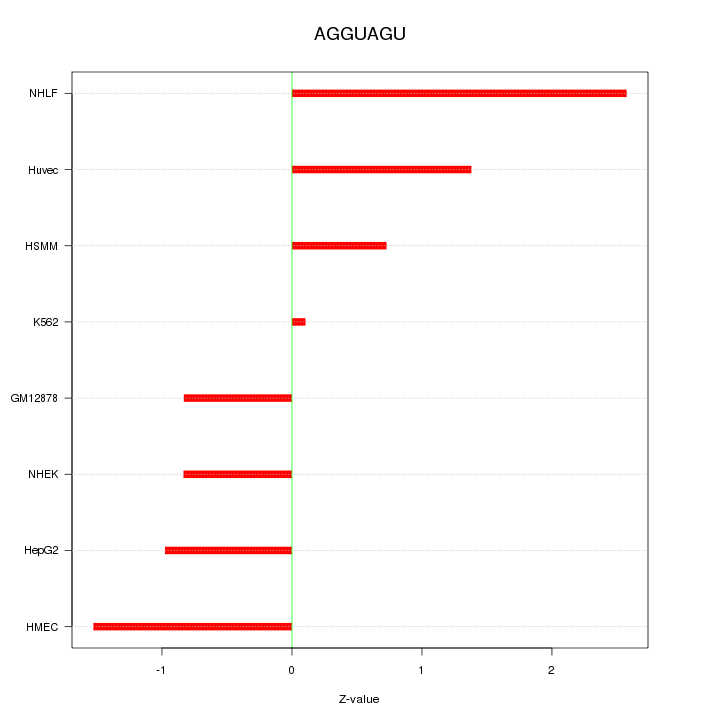
| 1.5 |
5.9 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.3 |
3.3 |
GO:0097435 |
extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.3 |
0.8 |
GO:0045656 |
negative regulation of monocyte differentiation(GO:0045656) |
| 0.2 |
1.3 |
GO:0009082 |
branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.2 |
0.9 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.2 |
0.6 |
GO:0060318 |
definitive erythrocyte differentiation(GO:0060318) |
| 0.2 |
2.0 |
GO:0007351 |
tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.2 |
1.7 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.2 |
0.6 |
GO:0060574 |
trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 0.2 |
0.5 |
GO:0071635 |
intestinal epithelial cell differentiation(GO:0060575) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 |
0.6 |
GO:0070669 |
response to interleukin-2(GO:0070669) |
| 0.1 |
0.5 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.1 |
0.4 |
GO:0006930 |
substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 |
0.2 |
GO:0060394 |
obsolete positive regulation of S phase of mitotic cell cycle(GO:0045750) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 |
0.2 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 |
0.8 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 |
0.1 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.0 |
0.1 |
GO:0009134 |
nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 |
0.4 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 |
0.2 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.0 |
0.2 |
GO:1903019 |
negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.0 |
0.2 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.2 |
GO:0097576 |
vacuole fusion(GO:0097576) |
| 0.0 |
0.6 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.2 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.0 |
0.4 |
GO:0048260 |
positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 |
0.2 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 |
0.1 |
GO:0071428 |
rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 |
0.2 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 |
1.3 |
GO:0048704 |
embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 |
0.2 |
GO:0046051 |
GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 |
0.1 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.0 |
0.2 |
GO:0033523 |
histone monoubiquitination(GO:0010390) histone H2B ubiquitination(GO:0033523) |
| 0.0 |
0.4 |
GO:0050807 |
regulation of synapse organization(GO:0050807) |
| 0.0 |
1.0 |
GO:0010977 |
negative regulation of neuron projection development(GO:0010977) |
| 0.0 |
0.1 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.0 |
0.1 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.9 |
GO:0034332 |
adherens junction organization(GO:0034332) |