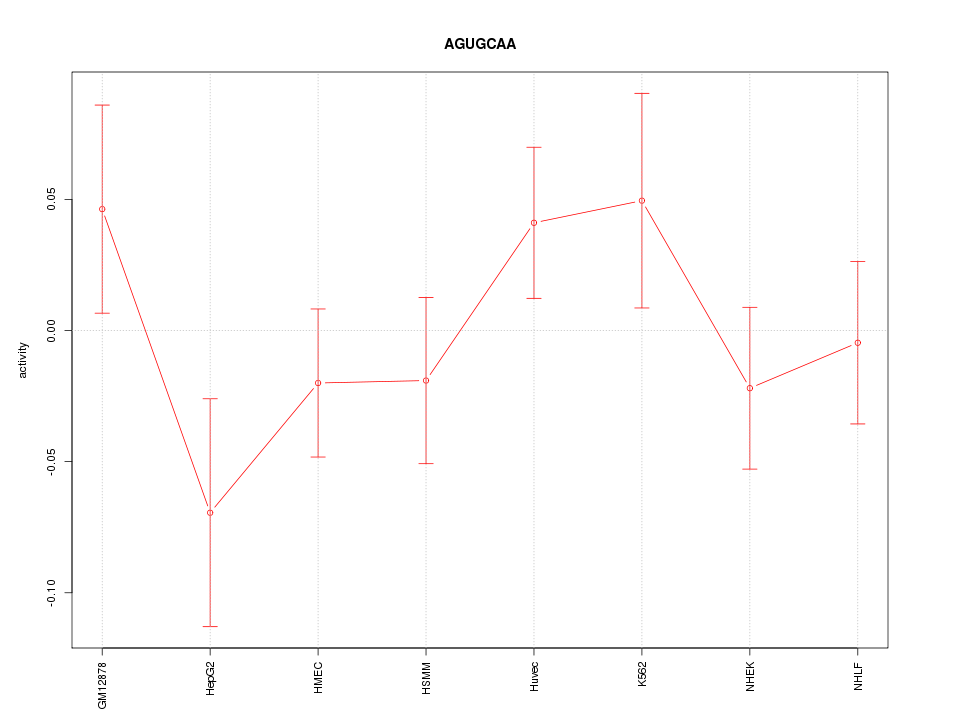
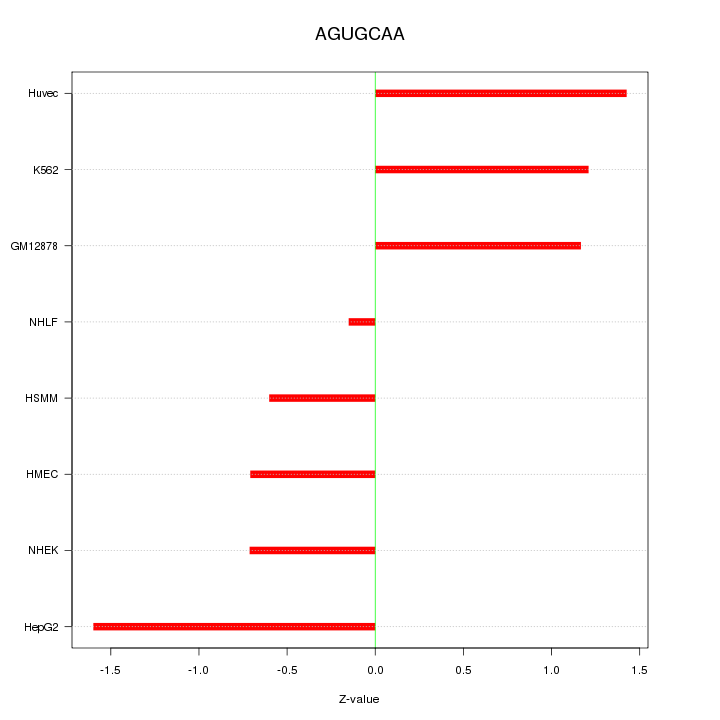
Motif ID: AGUGCAA
Z-value: 1.050

Mature miRNA associated with seed AGUGCAA:
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 0.7 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.2 | 0.6 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 0.4 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.2 | 0.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 0.3 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.2 | 0.5 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.9 | GO:0035630 | glycolate metabolic process(GO:0009441) bone mineralization involved in bone maturation(GO:0035630) |
| 0.1 | 0.4 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.1 | 0.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.5 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.7 | GO:0072205 | collecting duct development(GO:0072044) metanephric tubule development(GO:0072170) metanephric collecting duct development(GO:0072205) metanephric epithelium development(GO:0072207) metanephric nephron tubule development(GO:0072234) metanephric nephron epithelium development(GO:0072243) |
| 0.1 | 0.7 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.1 | 0.4 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.6 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 0.5 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.7 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.3 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.1 | 0.5 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 0.3 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.1 | 0.6 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.5 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 | 0.4 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 0.2 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 0.4 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) positive regulation of dopamine receptor signaling pathway(GO:0060161) caveola assembly(GO:0070836) membrane assembly(GO:0071709) |
| 0.1 | 1.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.1 | GO:0071230 | positive regulation of TOR signaling(GO:0032008) cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 0.3 | GO:0002328 | lymphoid progenitor cell differentiation(GO:0002320) pro-B cell differentiation(GO:0002328) |
| 0.1 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.4 | GO:0032916 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.1 | 0.6 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.0 | 0.7 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.6 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.3 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 1.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell differentiation(GO:0033078) extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.1 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.5 | GO:0009220 | pyrimidine ribonucleotide biosynthetic process(GO:0009220) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) snRNA transport(GO:0051030) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.3 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.6 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:0043921 | modulation by host of viral transcription(GO:0043921) modulation by host of symbiont transcription(GO:0052472) |
| 0.0 | 0.4 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.4 | GO:0033523 | histone monoubiquitination(GO:0010390) histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.2 | GO:0045651 | monocyte activation(GO:0042117) positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0060674 | placenta blood vessel development(GO:0060674) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.4 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.5 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 1.0 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.0 | 0.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.6 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 1.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.4 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.2 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.6 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.0 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.1 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.9 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 1.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 2.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 3.3 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.7 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.7 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.1 | 1.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.1 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.1 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.2 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 0.4 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.2 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.1 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.7 | GO:0016896 | exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 1.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0016174 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0004428 | obsolete inositol or phosphatidylinositol kinase activity(GO:0004428) |
| 0.0 | 0.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 2.0 | GO:0016566 | obsolete specific transcriptional repressor activity(GO:0016566) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.5 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.7 | GO:0003704 | obsolete specific RNA polymerase II transcription factor activity(GO:0003704) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 2.2 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.0 | 0.0 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.1 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 1.6 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | ST_STAT3_PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |