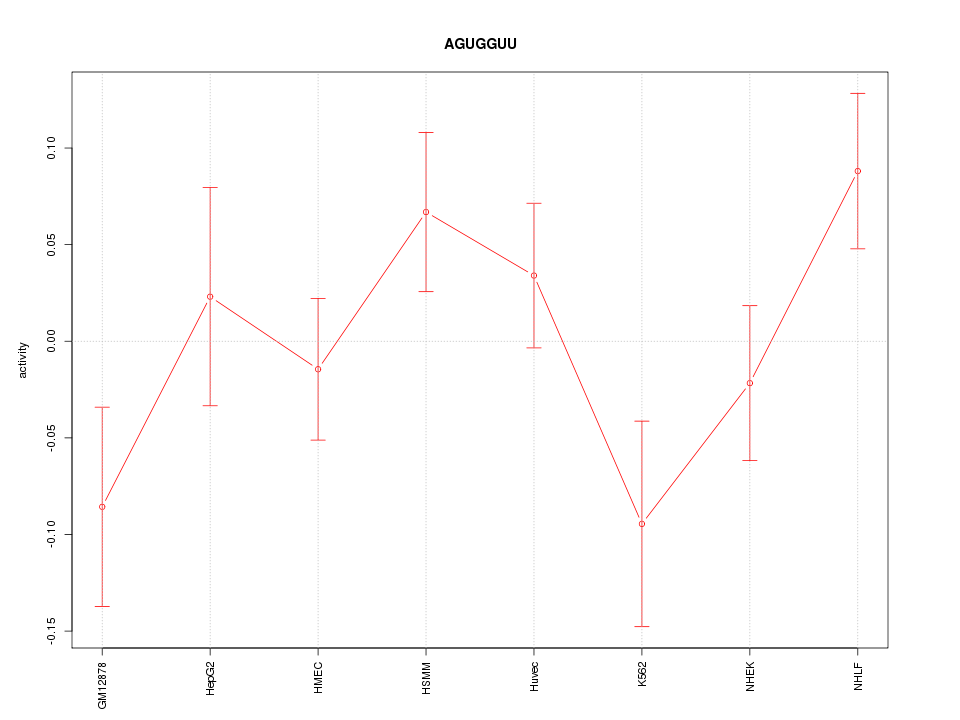
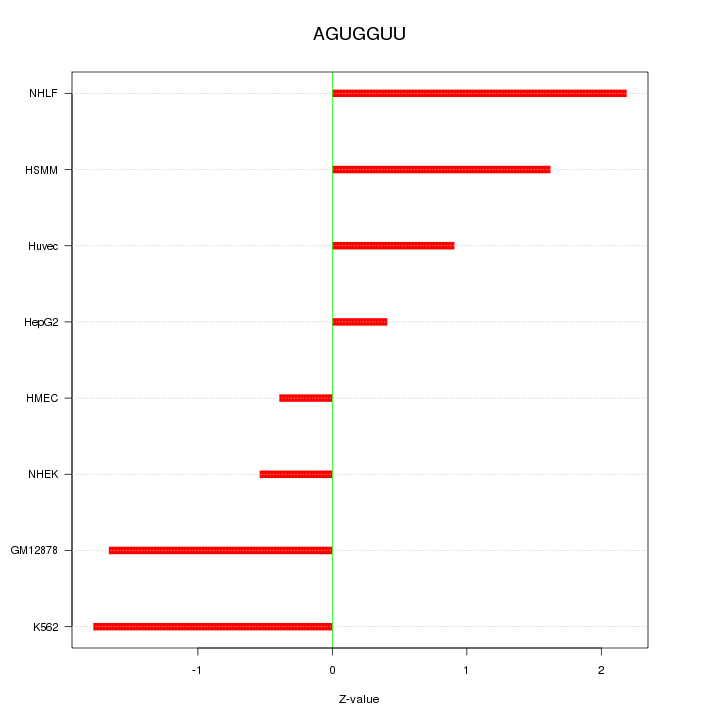
Motif ID: AGUGGUU
Z-value: 1.359

Mature miRNA associated with seed AGUGGUU:
| Name | miRBase Accession |
|---|---|
| hsa-miR-140-5p | MIMAT0000431 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.3 | 3.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.3 | 0.8 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.2 | 0.9 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.2 | 1.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 1.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 1.6 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.2 | 1.6 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.2 | 0.6 | GO:0060729 | negative regulation of interleukin-23 production(GO:0032707) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.8 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.6 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 1.6 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.2 | GO:0030812 | muscle hypertrophy in response to stress(GO:0003299) response to stimulus involved in regulation of muscle adaptation(GO:0014874) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.1 | 0.4 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.3 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 1.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.3 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.5 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0060129 | corticotropin hormone secreting cell differentiation(GO:0060128) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.4 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 1.0 | GO:0009067 | aspartate family amino acid biosynthetic process(GO:0009067) |
| 0.0 | 0.3 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 1.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.2 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 | 0.2 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.2 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 1.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.8 | GO:0008064 | regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0090174 | organelle membrane fusion(GO:0090174) |
| 0.0 | 0.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.0 | GO:0009408 | response to heat(GO:0009408) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.2 | 3.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.2 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.4 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.6 | GO:0019717 | obsolete synaptosome(GO:0019717) |
| 0.0 | 2.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.3 | 1.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.6 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 1.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.2 | 1.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 1.6 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 0.9 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 1.6 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 0.3 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 1.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 2.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | SIG_CHEMOTAXIS | Genes related to chemotaxis |