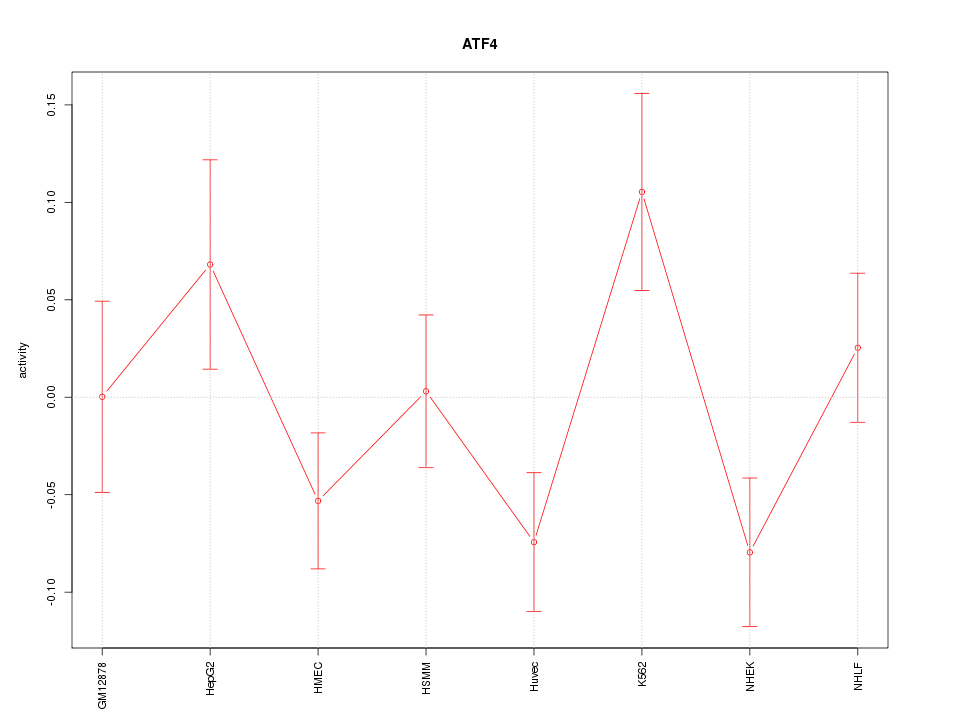
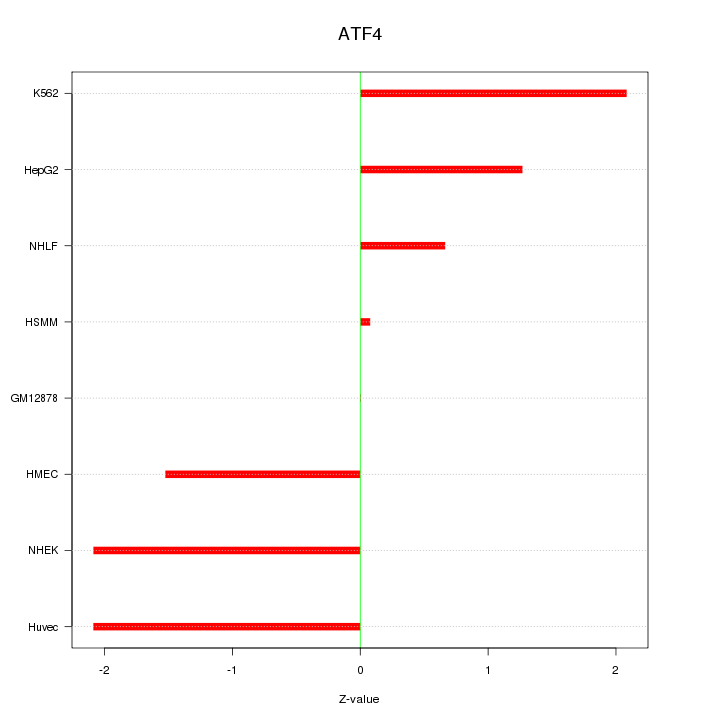
Motif ID: ATF4
Z-value: 1.476
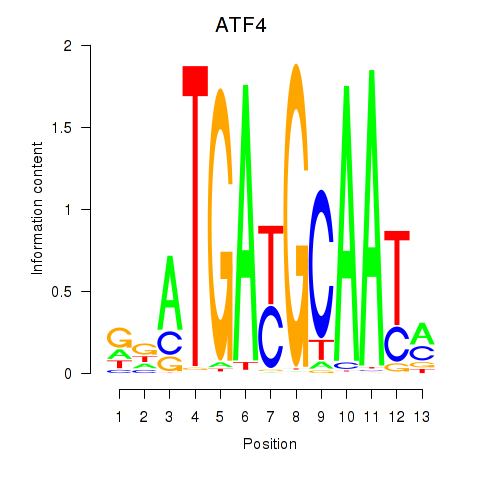
Transcription factors associated with ATF4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ATF4 | ENSG00000128272.10 | ATF4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:0007614 | short-term memory(GO:0007614) |
| 1.1 | 3.4 | GO:0043418 | cysteine biosynthetic process from serine(GO:0006535) homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) transsulfuration(GO:0019346) homocysteine catabolic process(GO:0043418) |
| 0.6 | 3.9 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.5 | 4.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.4 | 1.3 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.3 | 1.0 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.3 | 0.8 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.3 | 0.8 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 2.5 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 0.6 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 | 0.6 | GO:0097237 | response to cobalt ion(GO:0032025) cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.2 | 1.1 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.2 | 0.7 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.2 | 0.3 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 2.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.3 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 0.3 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 | 0.4 | GO:2000858 | renin-angiotensin regulation of aldosterone production(GO:0002018) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) positive regulation of NAD(P)H oxidase activity(GO:0033864) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.1 | 1.5 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.2 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) |
| 0.1 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.3 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.6 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 1.8 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 6.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 8.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 7.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.9 | 4.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.8 | 2.5 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.6 | 3.9 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.3 | 0.8 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.3 | 1.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 0.8 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 0.6 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 1.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.2 | 0.6 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 2.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 2.3 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.3 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.1 | 6.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 2.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.6 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |