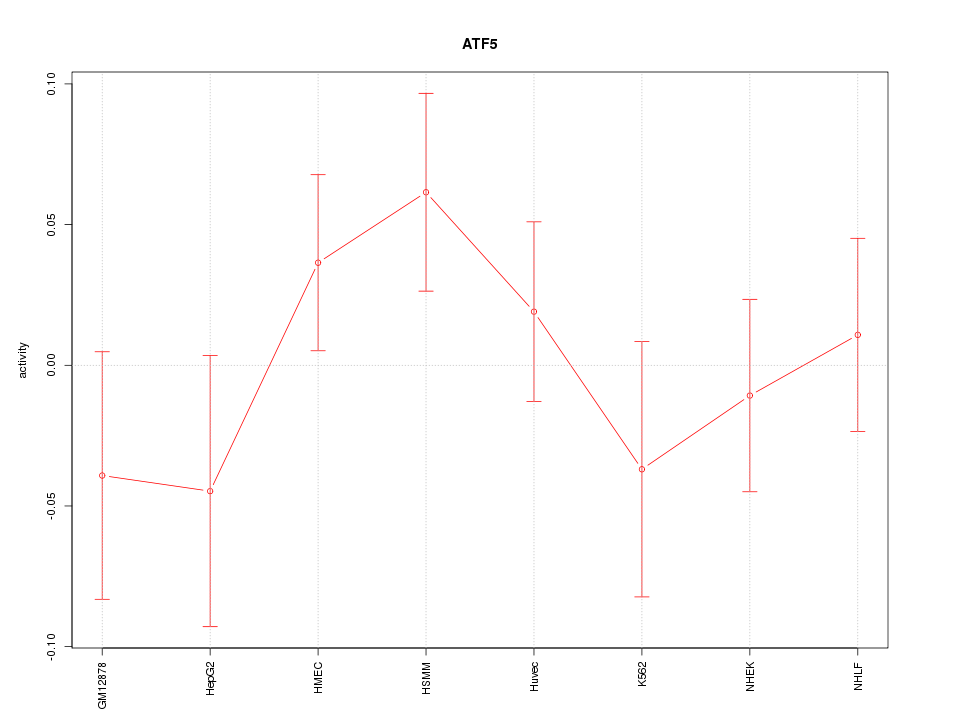
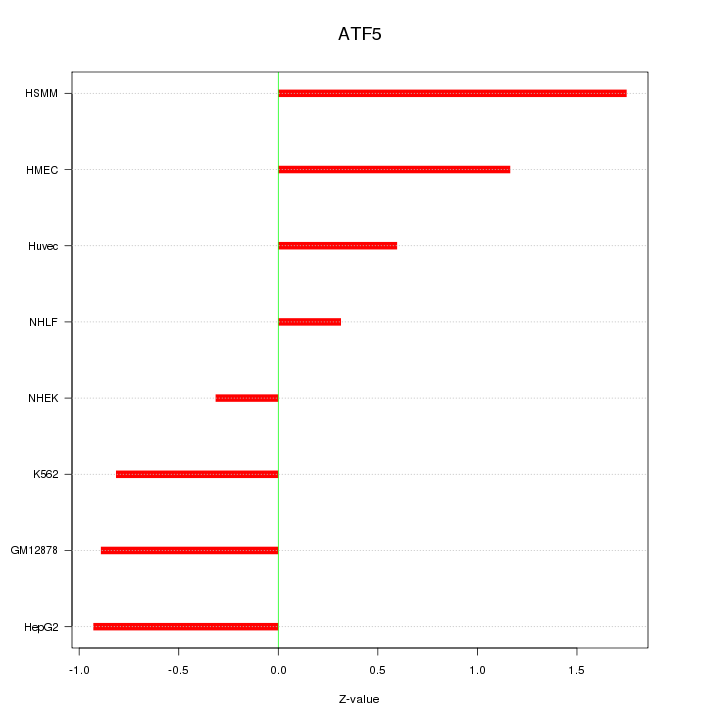
Motif ID: ATF5
Z-value: 0.955
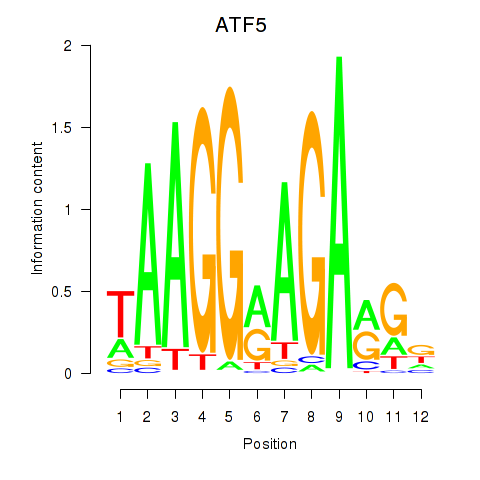
Transcription factors associated with ATF5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ATF5 | ENSG00000169136.4 | ATF5 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0060279 | progesterone secretion(GO:0042701) regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.6 | 1.8 | GO:0014889 | muscle atrophy(GO:0014889) regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.2 | 0.5 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.2 | 1.0 | GO:2000124 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 1.4 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.1 | 1.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 2.4 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 1.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 3.0 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 0.4 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.7 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.5 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 3.8 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.2 | GO:0010193 | response to ozone(GO:0010193) response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.8 | GO:1902749 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.6 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.3 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.3 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.4 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 2.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.9 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.4 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005624 | obsolete membrane fraction(GO:0005624) |
| 0.0 | 0.6 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.7 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0048184 | obsolete follistatin binding(GO:0048184) |
| 0.3 | 1.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 1.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 2.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 3.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |