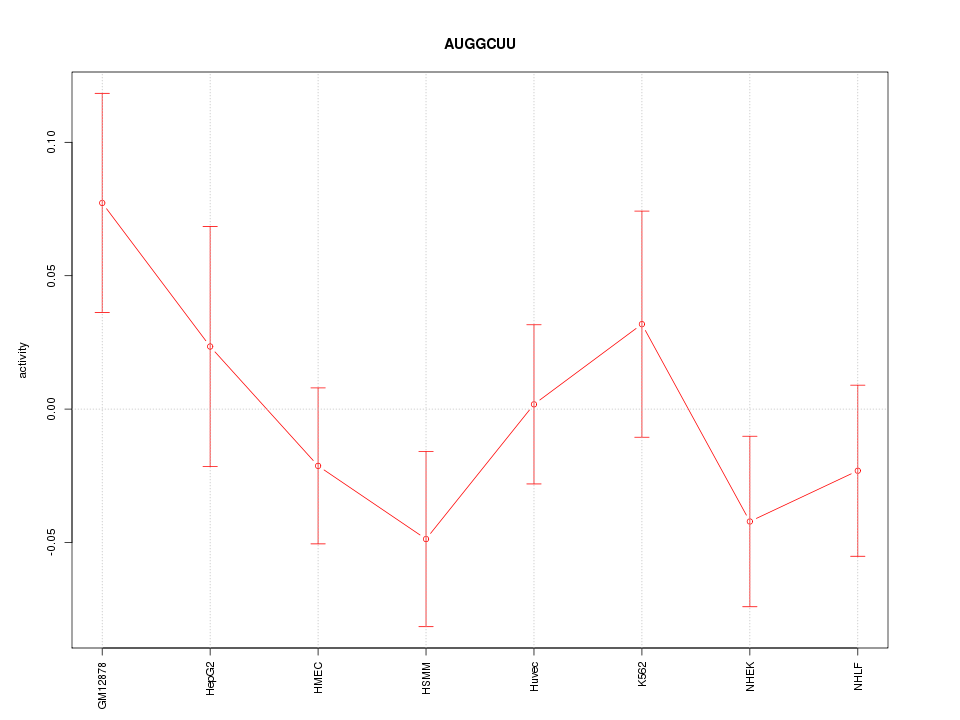
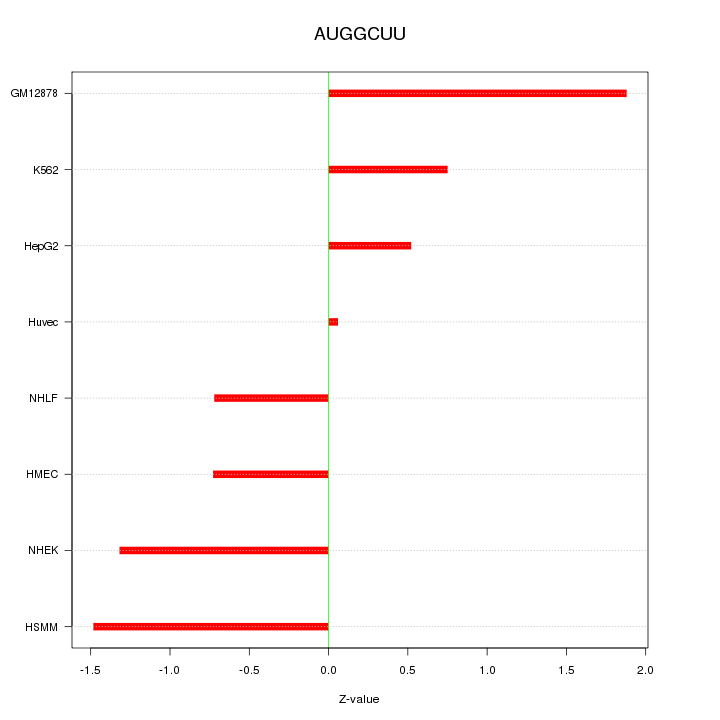
Motif ID: AUGGCUU
Z-value: 1.082

Mature miRNA associated with seed AUGGCUU:
| Name | miRBase Accession |
|---|---|
| hsa-miR-135a-5p | MIMAT0000428 |
| hsa-miR-135b-5p | MIMAT0000758 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.2 | 1.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.2 | 1.3 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.1 | 0.6 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 0.4 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.6 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.7 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.6 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.1 | 1.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.3 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.3 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.1 | 0.2 | GO:0060061 | regulation of collateral sprouting(GO:0048670) Spemann organizer formation(GO:0060061) |
| 0.1 | 0.6 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.5 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.1 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.2 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.1 | 1.0 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.2 | GO:0007262 | STAT protein import into nucleus(GO:0007262) regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 | 0.3 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 0.2 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.2 | GO:0014889 | muscle atrophy(GO:0014889) regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.4 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.1 | 0.3 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0016242 | regulation of macroautophagy(GO:0016241) negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.3 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.0 | 0.7 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.3 | GO:0051895 | negative regulation of stress fiber assembly(GO:0051497) negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0021896 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.3 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.4 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0045350 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.3 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0035751 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.2 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.2 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.3 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.3 | GO:0034339 | obsolete regulation of transcription from RNA polymerase II promoter by nuclear hormone receptor(GO:0034339) |
| 0.0 | 1.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.3 | GO:0045823 | positive regulation of heart contraction(GO:0045823) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.2 | GO:0075713 | establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0010921 | regulation of phosphatase activity(GO:0010921) regulation of protein dephosphorylation(GO:0035304) regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) synaptic cleft(GO:0043083) |
| 0.1 | 0.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.3 | GO:0042405 | nuclear RNA export factor complex(GO:0042272) nuclear inclusion body(GO:0042405) |
| 0.1 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 1.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.4 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.5 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.6 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 1.3 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.2 | 0.6 | GO:0015247 | aminophospholipid transporter activity(GO:0015247) |
| 0.2 | 1.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 0.5 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.4 | GO:0034701 | tripeptidase activity(GO:0034701) |
| 0.1 | 1.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.2 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 1.1 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 0.3 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 1.0 | GO:0016896 | exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.1 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.1 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.2 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.3 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 1.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 1.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.5 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.4 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |