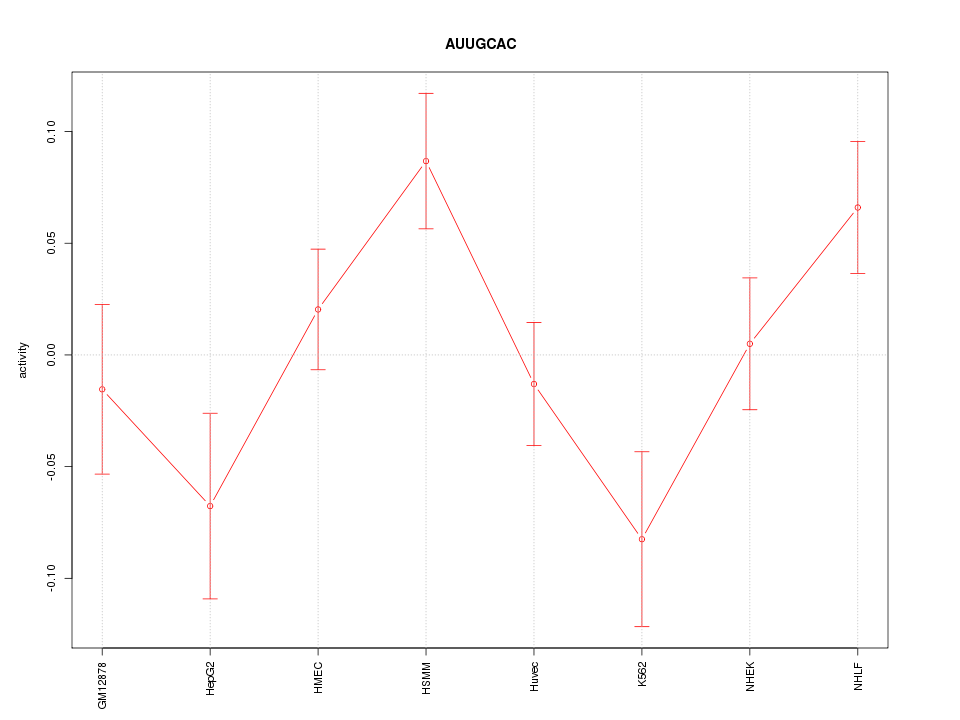
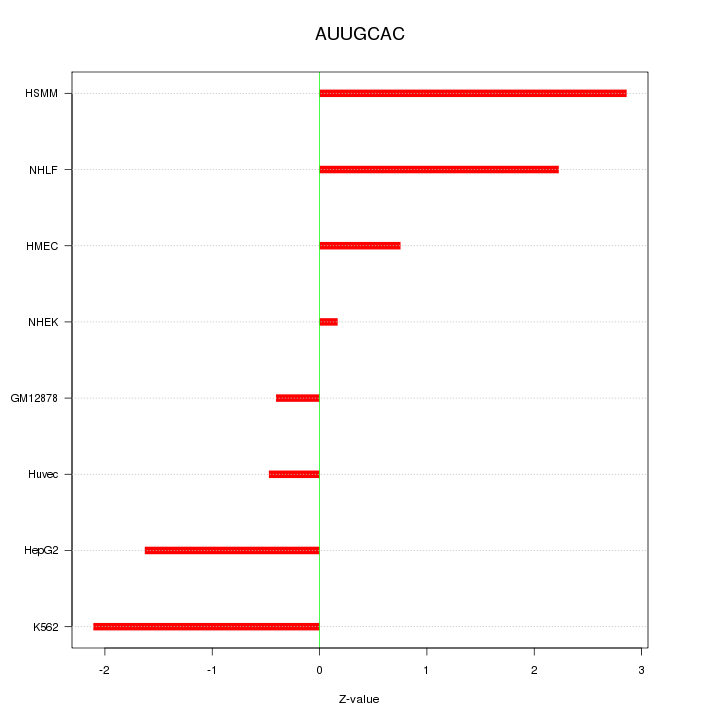
| 1.3 |
3.8 |
GO:2000064 |
regulation of cortisol biosynthetic process(GO:2000064) |
| 0.8 |
3.2 |
GO:0045112 |
integrin biosynthetic process(GO:0045112) |
| 0.5 |
2.5 |
GO:0060414 |
aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.5 |
2.8 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 |
2.5 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.4 |
2.7 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.3 |
1.0 |
GO:0031133 |
regulation of axon diameter(GO:0031133) regulation of cell projection size(GO:0032536) |
| 0.3 |
1.0 |
GO:0032510 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.3 |
1.0 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.3 |
2.9 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.3 |
1.3 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.3 |
1.9 |
GO:0042635 |
negative regulation of gonadotropin secretion(GO:0032277) positive regulation of hair cycle(GO:0042635) negative regulation of follicle-stimulating hormone secretion(GO:0046882) positive regulation of hair follicle development(GO:0051798) |
| 0.3 |
0.9 |
GO:0019367 |
fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.3 |
0.8 |
GO:0021836 |
chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.3 |
1.6 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 |
1.9 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.2 |
0.9 |
GO:0000212 |
meiotic spindle organization(GO:0000212) |
| 0.2 |
0.9 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) long-term synaptic potentiation(GO:0060291) |
| 0.2 |
1.4 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.2 |
0.7 |
GO:0010837 |
regulation of keratinocyte proliferation(GO:0010837) |
| 0.2 |
1.3 |
GO:0035313 |
wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.2 |
1.5 |
GO:0046349 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.2 |
0.8 |
GO:0045715 |
negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.2 |
1.6 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.2 |
0.2 |
GO:0043949 |
regulation of cAMP-mediated signaling(GO:0043949) |
| 0.2 |
0.9 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 |
0.6 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.1 |
2.4 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
1.5 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.1 |
0.4 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.1 |
0.9 |
GO:0031274 |
pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 |
0.6 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 |
3.7 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.1 |
0.6 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.1 |
2.1 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.1 |
0.3 |
GO:0009134 |
nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 |
1.1 |
GO:0021513 |
spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.1 |
0.3 |
GO:0031643 |
positive regulation of myelination(GO:0031643) |
| 0.1 |
0.4 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.1 |
0.6 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.7 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.3 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.1 |
0.2 |
GO:0042524 |
negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.1 |
1.4 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.1 |
0.4 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 |
0.6 |
GO:0044273 |
sulfur compound catabolic process(GO:0044273) |
| 0.1 |
1.0 |
GO:0070301 |
cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 |
0.2 |
GO:0034205 |
beta-amyloid formation(GO:0034205) |
| 0.1 |
1.1 |
GO:0032964 |
collagen biosynthetic process(GO:0032964) |
| 0.1 |
0.9 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.1 |
0.6 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 |
1.5 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.1 |
0.2 |
GO:0071428 |
rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 |
0.1 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 |
0.5 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.1 |
0.2 |
GO:0051409 |
response to nitrosative stress(GO:0051409) |
| 0.1 |
1.0 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.3 |
GO:0050685 |
positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 |
0.4 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.2 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
1.1 |
GO:0030838 |
positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 |
0.1 |
GO:0046639 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of alpha-beta T cell differentiation(GO:0046639) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.0 |
0.3 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 |
0.3 |
GO:0044060 |
regulation of endocrine process(GO:0044060) |
| 0.0 |
0.1 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 |
0.2 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.3 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
3.8 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.1 |
GO:0003159 |
endocardium development(GO:0003157) morphogenesis of an endothelium(GO:0003159) endocardium morphogenesis(GO:0003160) |
| 0.0 |
0.2 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 |
0.9 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.6 |
GO:0002040 |
sprouting angiogenesis(GO:0002040) |
| 0.0 |
0.7 |
GO:0090003 |
regulation of establishment of protein localization to plasma membrane(GO:0090003) regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 |
0.1 |
GO:0000768 |
syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 |
0.6 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.0 |
0.1 |
GO:0045046 |
protein import into peroxisome membrane(GO:0045046) |
| 0.0 |
1.9 |
GO:0031668 |
cellular response to extracellular stimulus(GO:0031668) |
| 0.0 |
1.0 |
GO:0023014 |
MAPK cascade(GO:0000165) signal transduction by protein phosphorylation(GO:0023014) |
| 0.0 |
0.2 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 |
0.4 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 |
0.3 |
GO:0007184 |
SMAD protein import into nucleus(GO:0007184) |
| 0.0 |
0.6 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 |
0.3 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.9 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.0 |
1.3 |
GO:0010977 |
negative regulation of neuron projection development(GO:0010977) |
| 0.0 |
1.6 |
GO:0071772 |
BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 |
0.3 |
GO:0015884 |
folic acid transport(GO:0015884) |
| 0.0 |
0.1 |
GO:0002328 |
pro-B cell differentiation(GO:0002328) |
| 0.0 |
0.3 |
GO:0045730 |
respiratory burst(GO:0045730) |
| 0.0 |
0.8 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
0.1 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 |
1.7 |
GO:0030048 |
actin filament-based movement(GO:0030048) |
| 0.0 |
0.3 |
GO:0006024 |
glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 |
0.7 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.1 |
GO:0006549 |
isoleucine metabolic process(GO:0006549) |
| 0.0 |
0.0 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.1 |
GO:0007143 |
female meiotic division(GO:0007143) |
| 0.0 |
0.1 |
GO:0090224 |
regulation of spindle organization(GO:0090224) |
| 0.0 |
0.3 |
GO:0036503 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 |
0.0 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.7 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.0 |
0.6 |
GO:0006383 |
transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 |
0.0 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 |
0.0 |
GO:0007196 |
adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
1.6 |
GO:0043087 |
regulation of GTPase activity(GO:0043087) |