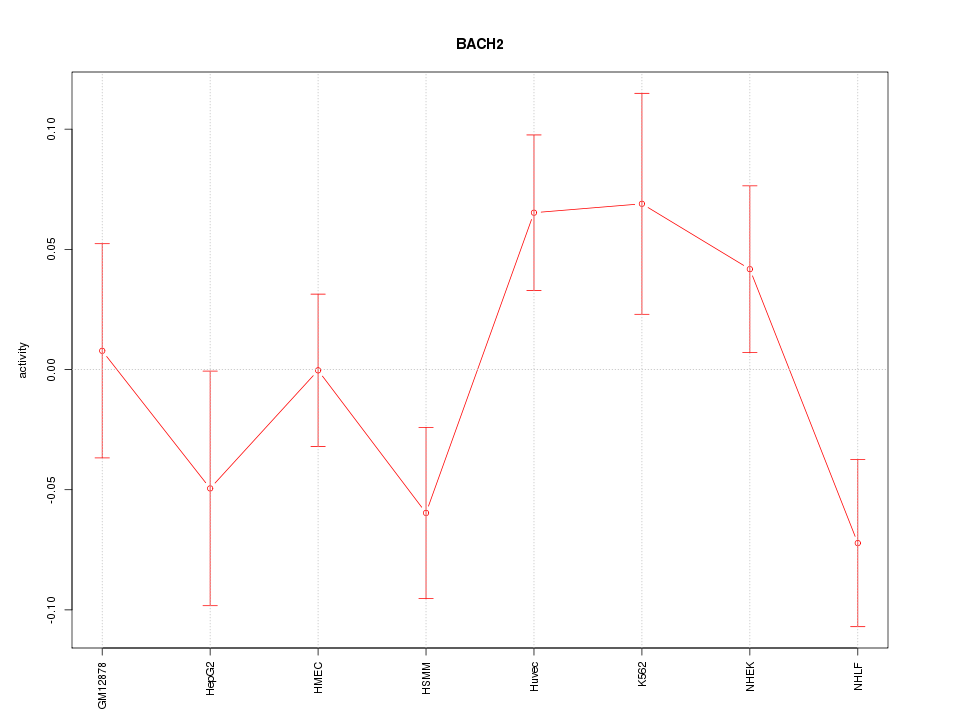
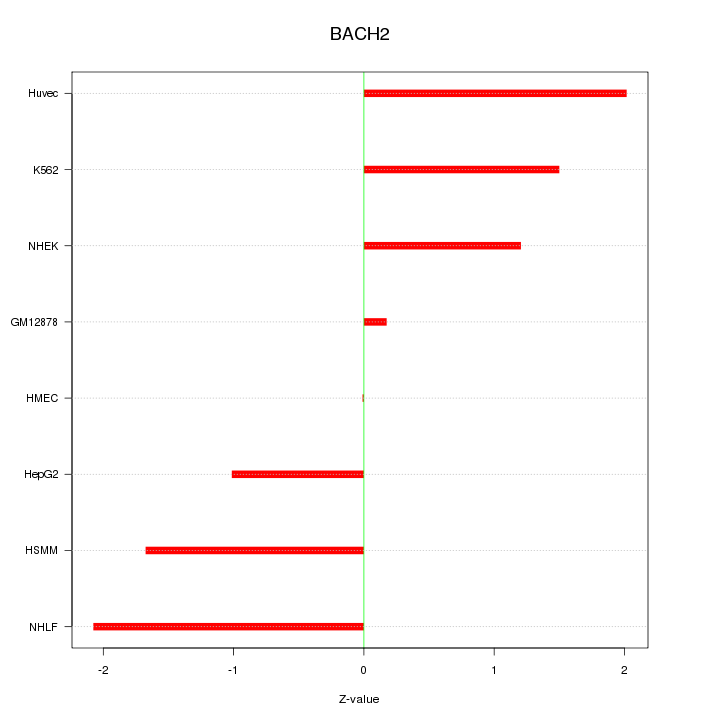
Motif ID: BACH2
Z-value: 1.412
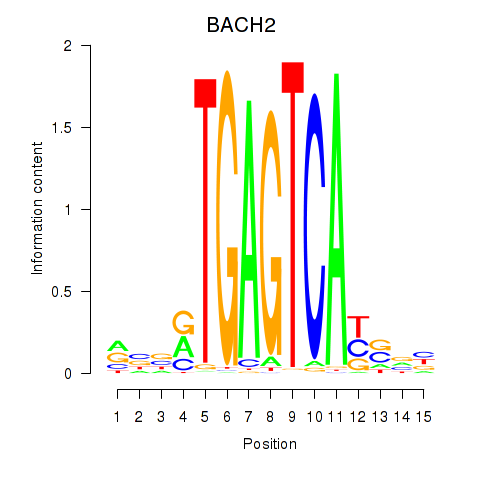
Transcription factors associated with BACH2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| BACH2 | ENSG00000112182.10 | BACH2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.4 | 1.3 | GO:0010430 | ethanol catabolic process(GO:0006068) fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 1.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 1.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 1.1 | GO:0042325 | regulation of phosphorylation(GO:0042325) |
| 0.2 | 0.7 | GO:0007262 | STAT protein import into nucleus(GO:0007262) regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.2 | 4.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.6 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.1 | 1.4 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.5 | GO:0045938 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 | 0.8 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.9 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.1 | 0.6 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.4 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.4 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.0 | GO:0019359 | nicotinamide nucleotide biosynthetic process(GO:0019359) |
| 0.1 | 0.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.6 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.4 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.1 | 0.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.8 | GO:0010224 | response to UV-B(GO:0010224) |
| 0.1 | 0.8 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 2.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.2 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.1 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.7 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 1.1 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 1.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 1.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.4 | GO:0050684 | regulation of mRNA processing(GO:0050684) regulation of mRNA metabolic process(GO:1903311) |
| 0.0 | 0.9 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 0.2 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.6 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.8 | GO:0072164 | ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 0.4 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 2.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.4 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.8 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 1.8 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.8 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.5 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.0 | GO:0071280 | epithelial fluid transport(GO:0042045) cellular response to copper ion(GO:0071280) |
| 0.0 | 0.1 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.1 | GO:0061025 | membrane fusion(GO:0061025) |
| 0.0 | 1.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.1 | GO:0030147 | obsolete natriuresis(GO:0030147) |
| 0.0 | 0.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.4 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 1.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.2 | 1.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 1.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 2.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 4.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.5 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 2.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0033655 | host(GO:0018995) host cell cytoplasm(GO:0030430) host cell part(GO:0033643) host intracellular part(GO:0033646) host cell cytoplasm part(GO:0033655) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.4 | 4.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 1.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) exoribonuclease II activity(GO:0008859) |
| 0.4 | 1.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 1.0 | GO:0004473 | malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.5 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) |
| 0.1 | 1.0 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.1 | 2.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.7 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 1.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.5 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.8 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.1 | 1.2 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.1 | 1.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 1.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 3.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |