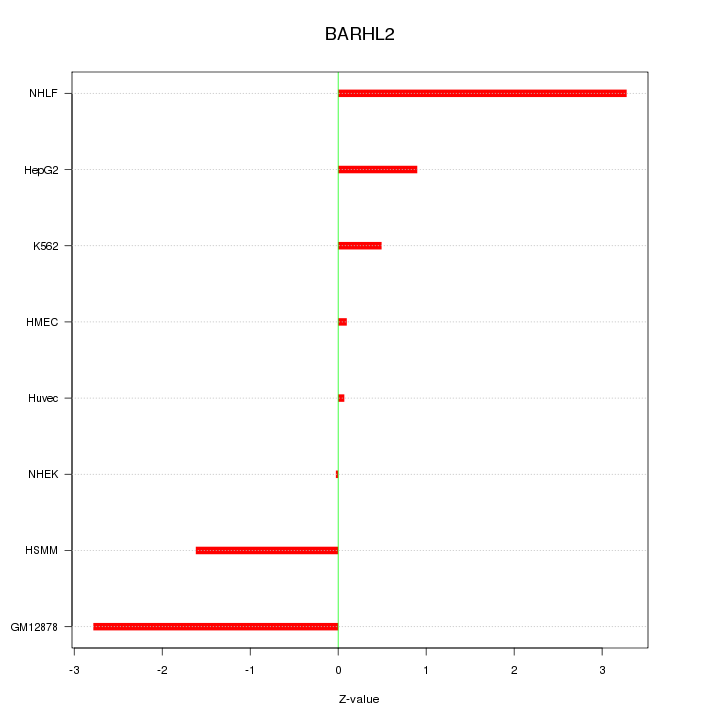
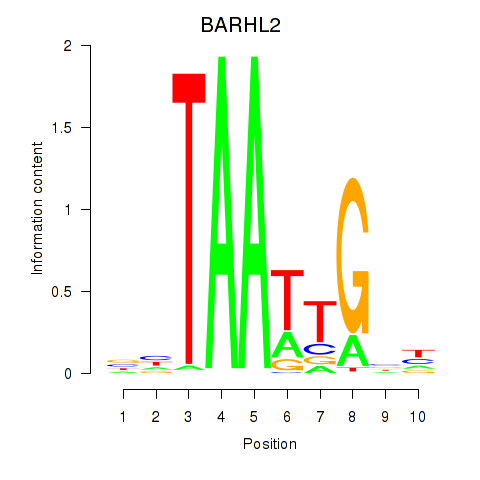
Motif ID: BARHL2
Z-value: 1.665
Transcription factors associated with BARHL2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| BARHL2 | ENSG00000143032.7 | BARHL2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 9.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.6 | 4.6 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.5 | 1.6 | GO:0035987 | endodermal cell fate commitment(GO:0001711) endodermal cell differentiation(GO:0035987) |
| 0.4 | 1.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.4 | 1.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.3 | 2.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.3 | 1.9 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.3 | 1.0 | GO:0014831 | intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.3 | 0.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.3 | 1.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.3 | 2.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.3 | 1.0 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.2 | 1.2 | GO:0035444 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) ferrous iron transport(GO:0015684) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) ferrous iron import(GO:0070627) iron ion import(GO:0097286) |
| 0.2 | 0.7 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.2 | 1.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 3.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 1.4 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 0.4 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.2 | 0.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 0.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 0.5 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 0.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 0.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.4 | GO:2000644 | low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.1 | 1.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.4 | GO:0090467 | response to herbicide(GO:0009635) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.1 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.1 | 0.6 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 0.3 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.1 | 1.0 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.7 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.8 | GO:0018126 | protein hydroxylation(GO:0018126) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.3 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.4 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.1 | 0.3 | GO:0060209 | estrus(GO:0060209) |
| 0.1 | 0.3 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.4 | GO:0034695 | response to prostaglandin E(GO:0034695) |
| 0.1 | 0.8 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.8 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 0.7 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.1 | GO:0072331 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) signal transduction by p53 class mediator(GO:0072331) intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.4 | GO:0019934 | cGMP-mediated signaling(GO:0019934) regulation of phagocytosis, engulfment(GO:0060099) positive regulation of phagocytosis, engulfment(GO:0060100) regulation of membrane invagination(GO:1905153) positive regulation of membrane invagination(GO:1905155) |
| 0.0 | 0.4 | GO:0060678 | dichotomous subdivision of terminal units involved in ureteric bud branching(GO:0060678) |
| 0.0 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 1.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.3 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 1.5 | GO:0050931 | melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 | 0.3 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.0 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.7 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 2.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 0.3 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.9 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:1904742 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) regulation of telomeric DNA binding(GO:1904742) positive regulation of telomeric DNA binding(GO:1904744) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.3 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 0.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 1.0 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.8 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of wound healing(GO:0061045) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0030324 | lung development(GO:0030324) |
| 0.0 | 0.1 | GO:1903301 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.2 | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 5.6 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.1 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.3 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.9 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.2 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0030449 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.5 | GO:0030866 | cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.1 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.3 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.1 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.1 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.6 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 0.4 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.3 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.1 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 | 0.0 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.1 | GO:0022602 | ovulation cycle process(GO:0022602) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 16.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.4 | 1.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.2 | 1.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 0.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.5 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 0.3 | GO:0036452 | ESCRT III complex(GO:0000815) ESCRT complex(GO:0036452) |
| 0.1 | 1.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 1.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.2 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.3 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) synaptic cleft(GO:0043083) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.1 | GO:0042597 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.6 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 1.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 14.5 | GO:0044421 | extracellular region part(GO:0044421) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 0.9 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.4 | 2.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.4 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 1.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 1.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 1.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 1.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 1.2 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) ferrous iron transmembrane transporter activity(GO:0015093) lead ion transmembrane transporter activity(GO:0015094) nickel cation transmembrane transporter activity(GO:0015099) vanadium ion transmembrane transporter activity(GO:0015100) chromium ion transmembrane transporter activity(GO:0070835) |
| 0.2 | 9.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.8 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.2 | 1.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.7 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.5 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 12.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 1.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 3.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 2.2 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.2 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.1 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.2 | GO:0047042 | androsterone dehydrogenase (B-specific) activity(GO:0047042) |
| 0.1 | 0.4 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) |
| 0.1 | 0.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0004461 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.1 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 1.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.8 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 2.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0001228 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 1.2 | GO:0003704 | obsolete specific RNA polymerase II transcription factor activity(GO:0003704) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.9 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.6 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.1 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |