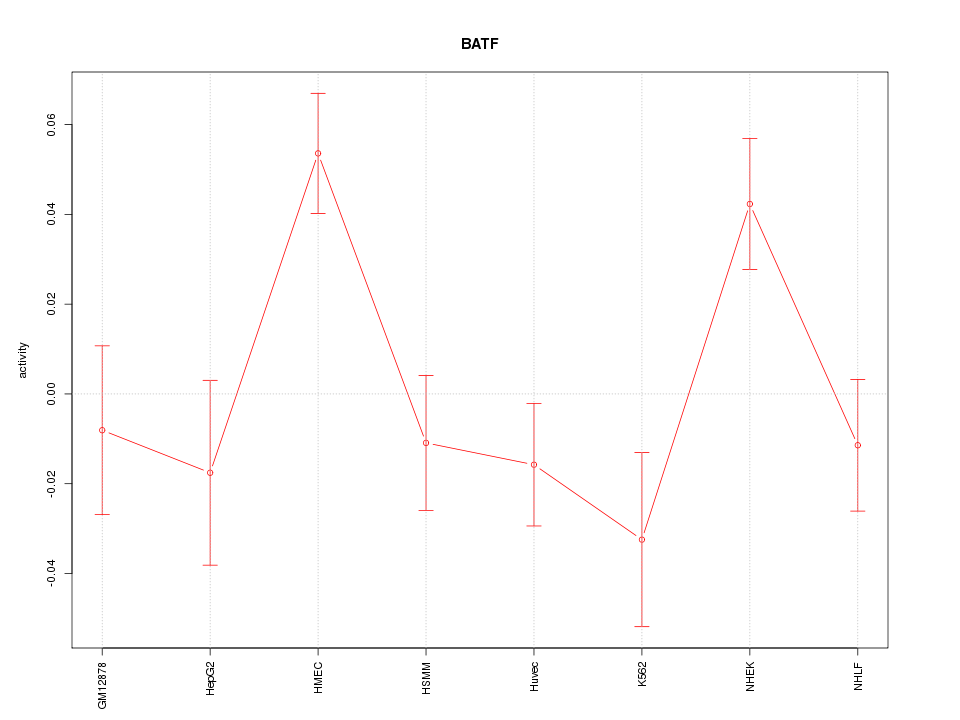
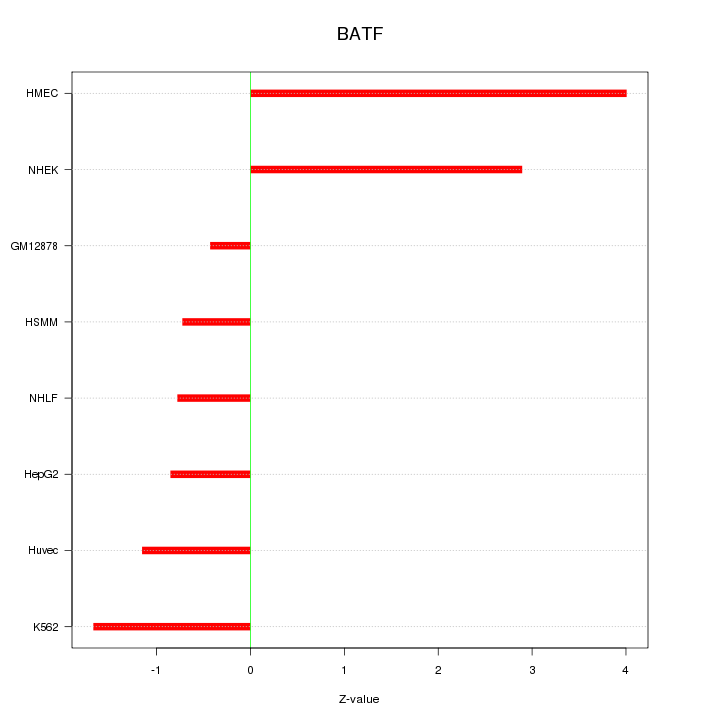
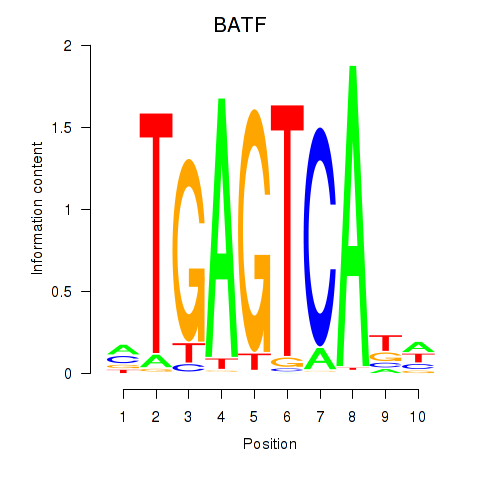
Motif ID: BATF
Z-value: 1.957
Transcription factors associated with BATF:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| BATF | ENSG00000156127.6 | BATF |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0010193 | response to ozone(GO:0010193) |
| 1.3 | 3.8 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 1.0 | 3.0 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.0 | 28.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.9 | 3.7 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.8 | 2.3 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.6 | 1.9 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.5 | 2.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.5 | 1.5 | GO:0009635 | response to herbicide(GO:0009635) |
| 0.4 | 1.3 | GO:0048627 | myoblast development(GO:0048627) |
| 0.4 | 1.5 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.4 | 1.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 1.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.3 | 1.4 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.3 | 2.2 | GO:0090025 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.3 | 1.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 1.0 | GO:0051503 | purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) |
| 0.2 | 1.5 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.2 | 1.6 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.2 | 1.4 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.2 | 0.7 | GO:0034205 | beta-amyloid formation(GO:0034205) |
| 0.2 | 1.3 | GO:2000124 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.9 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.2 | 1.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.2 | 6.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 1.4 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.2 | 1.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.7 | GO:0033986 | response to methanol(GO:0033986) response to chromate(GO:0046687) |
| 0.2 | 0.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 0.6 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 0.8 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 0.5 | GO:0046102 | nicotinamide riboside catabolic process(GO:0006738) inosine metabolic process(GO:0046102) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.2 | 4.7 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 0.2 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.2 | 0.5 | GO:0060340 | pericardium morphogenesis(GO:0003344) optic cup formation involved in camera-type eye development(GO:0003408) diencephalon morphogenesis(GO:0048852) positive regulation of type I interferon-mediated signaling pathway(GO:0060340) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.1 | 0.6 | GO:0031223 | mechanosensory behavior(GO:0007638) response to auditory stimulus(GO:0010996) auditory behavior(GO:0031223) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 2.8 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 0.4 | GO:0016081 | synaptic vesicle docking(GO:0016081) positive regulation of neurotransmitter transport(GO:0051590) |
| 0.1 | 3.9 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.1 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 0.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 15.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.5 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 0.4 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 1.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.7 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.5 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 0.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.4 | GO:0071418 | uroporphyrinogen III biosynthetic process(GO:0006780) cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 1.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.4 | GO:0051170 | nuclear import(GO:0051170) |
| 0.1 | 0.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.2 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.1 | 0.5 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 2.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.1 | GO:0060462 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.1 | 0.2 | GO:1904742 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) regulation of telomeric DNA binding(GO:1904742) positive regulation of telomeric DNA binding(GO:1904744) |
| 0.1 | 0.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.3 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.1 | 0.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.6 | GO:0019359 | nicotinamide nucleotide biosynthetic process(GO:0019359) |
| 0.1 | 0.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 3.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.5 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.3 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.3 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.1 | 0.2 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.1 | 0.2 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.5 | GO:0060119 | auditory receptor cell development(GO:0060117) inner ear receptor cell development(GO:0060119) |
| 0.1 | 0.3 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.7 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.1 | GO:0097300 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.0 | GO:0034014 | response to triglyceride(GO:0034014) |
| 0.0 | 0.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 1.0 | GO:0060749 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.0 | 0.3 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 8.0 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 1.1 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.2 | GO:0060346 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0046132 | pyrimidine ribonucleoside biosynthetic process(GO:0046132) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 1.0 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.1 | GO:0070365 | hepatocyte differentiation(GO:0070365) cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.3 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 0.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:0090085 | protein K29-linked ubiquitination(GO:0035519) regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 2.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.8 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.1 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 2.7 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.3 | GO:0034661 | ncRNA catabolic process(GO:0034661) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.1 | GO:1902414 | protein localization to adherens junction(GO:0071896) protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.8 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0034776 | negative regulation of sodium:proton antiporter activity(GO:0032416) response to histamine(GO:0034776) negative regulation of dopamine receptor signaling pathway(GO:0060160) negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.2 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0032527 | retrograde protein transport, ER to cytosol(GO:0030970) protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 | 0.2 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.4 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 1.7 | GO:0009206 | ATP biosynthetic process(GO:0006754) ribonucleoside triphosphate biosynthetic process(GO:0009201) purine ribonucleoside triphosphate biosynthetic process(GO:0009206) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.2 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 | 1.0 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 1.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.0 | GO:0007418 | ventral midline development(GO:0007418) |
| 0.0 | 1.4 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.0 | GO:0009750 | response to fructose(GO:0009750) |
| 0.0 | 0.1 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.3 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.6 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 1.0 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.3 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) |
| 0.0 | 0.1 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.0 | 0.1 | GO:2000648 | positive regulation of stem cell proliferation(GO:2000648) |
| 0.0 | 0.1 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.2 | 18.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.5 | 3.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 2.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 1.9 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.3 | 1.4 | GO:0032449 | CBM complex(GO:0032449) |
| 0.3 | 5.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 7.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 1.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 0.7 | GO:0014802 | terminal cisterna(GO:0014802) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 5.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.1 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.1 | 2.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.8 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.1 | 1.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 1.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.4 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 3.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.7 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.1 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.1 | GO:0070938 | actomyosin contractile ring(GO:0005826) contractile ring(GO:0070938) |
| 0.1 | 1.2 | GO:0043205 | fibril(GO:0043205) |
| 0.1 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.2 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0031975 | organelle envelope(GO:0031967) envelope(GO:0031975) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 1.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0019861 | obsolete flagellum(GO:0019861) |
| 0.0 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0019867 | outer membrane(GO:0019867) |
| 0.0 | 1.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.5 | GO:0070161 | anchoring junction(GO:0070161) |
| 0.0 | 0.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.9 | 3.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.7 | 3.4 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.6 | 2.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.5 | 6.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.5 | 2.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 0.7 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.2 | 1.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 19.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 3.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.6 | GO:0004473 | malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 0.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.6 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 2.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.0 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.3 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 1.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.5 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.4 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 1.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 1.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.4 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 3.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.1 | 8.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0017096 | acetylserotonin O-methyltransferase activity(GO:0017096) |
| 0.1 | 0.3 | GO:0047026 | androsterone dehydrogenase (A-specific) activity(GO:0047026) |
| 0.1 | 1.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 1.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.5 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 1.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0050833 | pyruvate secondary active transmembrane transporter activity(GO:0005477) pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 1.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 1.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 1.1 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 1.1 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 7.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.5 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.9 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 27.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 1.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0070891 | peptidoglycan receptor activity(GO:0016019) lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 2.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) |
| 0.0 | 0.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0050062 | long-chain-fatty-acyl-CoA reductase activity(GO:0050062) |
| 0.0 | 0.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.0 | GO:0016635 | succinate dehydrogenase activity(GO:0000104) oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0004461 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.0 | GO:0017129 | triglyceride binding(GO:0017129) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.7 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.0 | 0.3 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.1 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.0 | 0.3 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |