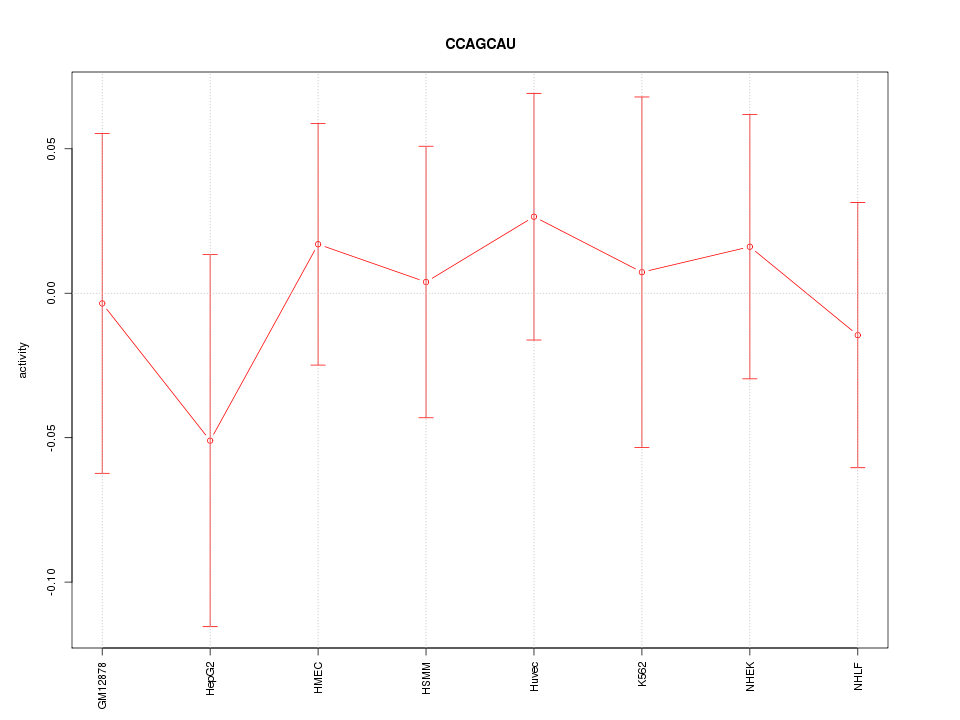
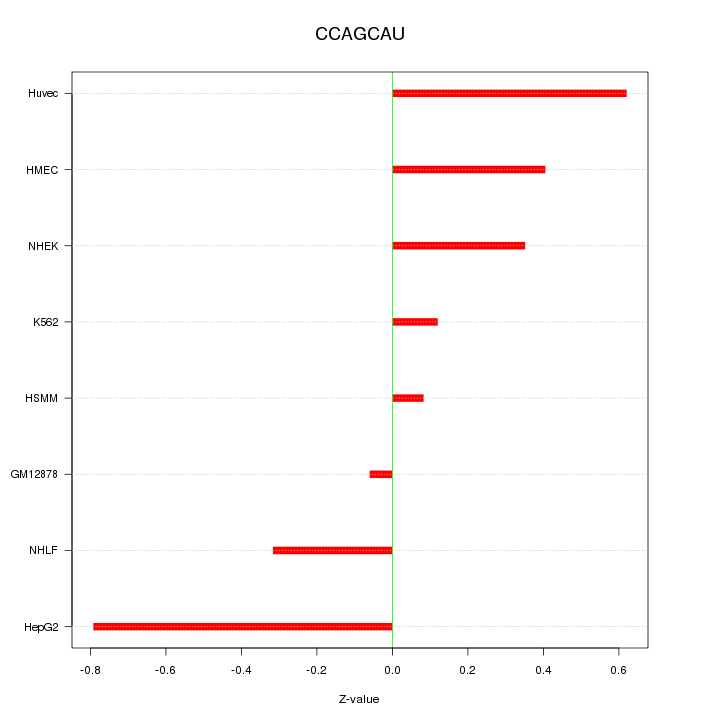
|
chr7_-_87849340
|
0.504
|
ENST00000419179.1
ENST00000265729.2
|
SRI
|
sorcin
|
|
chrX_+_64887512
|
0.464
|
ENST00000360270.5
|
MSN
|
moesin
|
|
chr15_+_39873268
|
0.385
|
ENST00000397591.2
ENST00000260356.5
|
THBS1
|
thrombospondin 1
|
|
chr18_-_53255766
|
0.264
|
ENST00000566286.1
ENST00000564999.1
ENST00000566279.1
ENST00000354452.3
ENST00000356073.4
|
TCF4
|
transcription factor 4
|
|
chr5_+_86564739
|
0.254
|
ENST00000456692.2
ENST00000512763.1
ENST00000506290.1
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr15_+_85359911
|
0.194
|
ENST00000258888.5
|
ALPK3
|
alpha-kinase 3
|
|
chr10_-_33623564
|
0.176
|
ENST00000374875.1
ENST00000374822.4
|
NRP1
|
neuropilin 1
|
|
chr15_+_92396920
|
0.170
|
ENST00000318445.6
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1
|
|
chr6_+_21593972
|
0.160
|
ENST00000244745.1
ENST00000543472.1
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr10_-_126849068
|
0.156
|
ENST00000494626.2
ENST00000337195.5
|
CTBP2
|
C-terminal binding protein 2
|
|
chr11_+_4116005
|
0.146
|
ENST00000300738.5
|
RRM1
|
ribonucleotide reductase M1
|
|
chr5_+_61602055
|
0.143
|
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A
|
|
chr11_+_113930291
|
0.121
|
ENST00000335953.4
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr3_-_9291063
|
0.120
|
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr6_+_16238786
|
0.111
|
ENST00000259727.4
|
GMPR
|
guanosine monophosphate reductase
|
|
chr1_-_154531095
|
0.107
|
ENST00000292211.4
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr3_-_134093395
|
0.106
|
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2
|
|
chr17_-_4269768
|
0.098
|
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1
|
|
chr4_-_55991752
|
0.096
|
ENST00000263923.4
|
KDR
|
kinase insert domain receptor (a type III receptor tyrosine kinase)
|
|
chr10_+_105726862
|
0.094
|
ENST00000335753.4
ENST00000369755.3
|
SLK
|
STE20-like kinase
|
|
chr3_+_118892362
|
0.093
|
ENST00000497685.1
ENST00000264234.3
|
UPK1B
|
uroplakin 1B
|
|
chr12_-_4647606
|
0.090
|
ENST00000261250.3
ENST00000541014.1
ENST00000545746.1
ENST00000542080.1
|
C12orf4
|
chromosome 12 open reading frame 4
|
|
chr22_-_22221900
|
0.089
|
ENST00000215832.6
ENST00000398822.3
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr5_-_107006596
|
0.086
|
ENST00000333274.6
|
EFNA5
|
ephrin-A5
|
|
chr15_-_75249793
|
0.083
|
ENST00000322177.5
|
RPP25
|
ribonuclease P/MRP 25kDa subunit
|
|
chr18_-_9614515
|
0.082
|
ENST00000400556.3
ENST00000400555.3
|
PPP4R1
|
protein phosphatase 4, regulatory subunit 1
|
|
chr7_+_98972298
|
0.081
|
ENST00000252725.5
|
ARPC1B
|
actin related protein 2/3 complex, subunit 1B, 41kDa
|
|
chr1_+_7831323
|
0.080
|
ENST00000054666.6
|
VAMP3
|
vesicle-associated membrane protein 3
|
|
chr11_-_10562710
|
0.080
|
ENST00000528665.1
ENST00000265981.2
|
RNF141
|
ring finger protein 141
|
|
chr5_-_126366500
|
0.078
|
ENST00000308660.5
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase
|
|
chr14_+_103243813
|
0.077
|
ENST00000560371.1
ENST00000347662.4
ENST00000392745.2
ENST00000539721.1
ENST00000560463.1
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr1_+_153606513
|
0.074
|
ENST00000368694.3
|
CHTOP
|
chromatin target of PRMT1
|
|
chr6_+_63921351
|
0.071
|
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C
|
|
chr3_-_176914238
|
0.067
|
ENST00000430069.1
ENST00000428970.1
|
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1
|
|
chr12_-_57400227
|
0.061
|
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr1_+_201798269
|
0.059
|
ENST00000361565.4
|
IPO9
|
importin 9
|
|
chr8_-_98290087
|
0.059
|
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5
|
|
chr3_+_100428268
|
0.058
|
ENST00000240851.4
|
TFG
|
TRK-fused gene
|
|
chr4_-_149365827
|
0.057
|
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr7_-_25019760
|
0.056
|
ENST00000352860.1
ENST00000353930.1
ENST00000431825.2
ENST00000313367.2
|
OSBPL3
|
oxysterol binding protein-like 3
|
|
chr1_+_154975110
|
0.054
|
ENST00000535420.1
ENST00000368426.3
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr12_+_62654119
|
0.054
|
ENST00000353364.3
ENST00000549523.1
ENST00000280377.5
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr15_+_45422131
|
0.054
|
ENST00000321429.4
|
DUOX1
|
dual oxidase 1
|
|
chrX_+_109246285
|
0.052
|
ENST00000372073.1
ENST00000372068.2
ENST00000288381.4
|
TMEM164
|
transmembrane protein 164
|
|
chr14_+_67826709
|
0.047
|
ENST00000256383.4
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr8_+_120079478
|
0.047
|
ENST00000332843.2
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr14_+_65171099
|
0.046
|
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr3_-_179754706
|
0.045
|
ENST00000465751.1
ENST00000467460.1
|
PEX5L
|
peroxisomal biogenesis factor 5-like
|
|
chr3_-_135914615
|
0.045
|
ENST00000309993.2
|
MSL2
|
male-specific lethal 2 homolog (Drosophila)
|
|
chr17_+_61086917
|
0.044
|
ENST00000424789.2
ENST00000389520.4
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr3_+_51422478
|
0.043
|
ENST00000528157.1
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr19_+_1752372
|
0.041
|
ENST00000382349.4
|
ONECUT3
|
one cut homeobox 3
|
|
chr20_-_39317868
|
0.041
|
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B
|
|
chr8_+_6565854
|
0.040
|
ENST00000285518.6
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5
|
|
chr7_+_139044621
|
0.040
|
ENST00000354926.4
|
C7orf55-LUC7L2
|
C7orf55-LUC7L2 readthrough
|
|
chr13_-_107220455
|
0.038
|
ENST00000400198.3
|
ARGLU1
|
arginine and glutamate rich 1
|
|
chr20_-_1373726
|
0.037
|
ENST00000400137.4
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chr17_+_30677136
|
0.037
|
ENST00000394670.4
ENST00000321233.6
ENST00000394673.2
ENST00000341711.6
ENST00000579634.1
ENST00000580759.1
ENST00000342555.6
ENST00000577908.1
ENST00000394679.5
ENST00000582165.1
|
ZNF207
|
zinc finger protein 207
|
|
chr4_+_41992489
|
0.036
|
ENST00000264451.7
|
SLC30A9
|
solute carrier family 30 (zinc transporter), member 9
|
|
chr4_+_39699664
|
0.035
|
ENST00000261427.5
ENST00000510934.1
ENST00000295963.6
|
UBE2K
|
ubiquitin-conjugating enzyme E2K
|
|
chr3_+_238273
|
0.035
|
ENST00000256509.2
|
CHL1
|
cell adhesion molecule L1-like
|
|
chr1_-_78148324
|
0.035
|
ENST00000370801.3
ENST00000433749.1
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr1_-_35658736
|
0.034
|
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr7_-_14942944
|
0.034
|
ENST00000403951.2
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr1_-_38273840
|
0.033
|
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing
|
|
chr9_-_80646374
|
0.033
|
ENST00000286548.4
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chrX_-_54522558
|
0.032
|
ENST00000375135.3
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr12_-_54982420
|
0.032
|
ENST00000257905.8
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A
|
|
chr7_+_77166592
|
0.031
|
ENST00000248594.6
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr20_-_41818373
|
0.031
|
ENST00000373187.1
ENST00000356100.2
ENST00000373184.1
ENST00000373190.1
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr10_-_75634260
|
0.031
|
ENST00000372765.1
ENST00000351293.3
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr16_+_68771128
|
0.031
|
ENST00000261769.5
ENST00000422392.2
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr12_-_10875831
|
0.031
|
ENST00000279550.7
ENST00000228251.4
|
YBX3
|
Y box binding protein 3
|
|
chr2_-_152684977
|
0.029
|
ENST00000428992.2
ENST00000295087.8
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr20_+_51588873
|
0.029
|
ENST00000371497.5
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr12_-_102874416
|
0.028
|
ENST00000392904.1
ENST00000337514.6
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr6_-_116601044
|
0.027
|
ENST00000368608.3
|
TSPYL1
|
TSPY-like 1
|
|
chr7_-_26240357
|
0.027
|
ENST00000354667.4
ENST00000356674.7
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr2_-_25475120
|
0.026
|
ENST00000380746.4
ENST00000402667.1
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr5_+_140560980
|
0.026
|
ENST00000361016.2
|
PCDHB16
|
protocadherin beta 16
|
|
chr5_+_141488070
|
0.026
|
ENST00000253814.4
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr10_-_105615164
|
0.025
|
ENST00000355946.2
ENST00000369774.4
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr1_+_244214577
|
0.025
|
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18
|
|
chr1_-_51763661
|
0.025
|
ENST00000530004.1
|
TTC39A
|
tetratricopeptide repeat domain 39A
|
|
chr4_-_168155730
|
0.025
|
ENST00000502330.1
ENST00000357154.3
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr14_-_76127519
|
0.024
|
ENST00000256319.6
|
C14orf1
|
chromosome 14 open reading frame 1
|
|
chr20_+_20348740
|
0.024
|
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1
|
|
chr16_-_79634595
|
0.024
|
ENST00000326043.4
ENST00000393350.1
|
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog
|
|
chr17_-_48474828
|
0.023
|
ENST00000576448.1
ENST00000225972.7
|
LRRC59
|
leucine rich repeat containing 59
|
|
chr2_+_210444142
|
0.023
|
ENST00000360351.4
ENST00000361559.4
|
MAP2
|
microtubule-associated protein 2
|
|
chr10_-_70287231
|
0.023
|
ENST00000609923.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16
|
|
chr2_+_48010221
|
0.023
|
ENST00000234420.5
|
MSH6
|
mutS homolog 6
|
|
chr5_+_43602750
|
0.018
|
ENST00000505678.2
ENST00000512422.1
ENST00000264663.5
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chrX_-_119445263
|
0.018
|
ENST00000309720.5
|
TMEM255A
|
transmembrane protein 255A
|
|
chr1_-_51425902
|
0.018
|
ENST00000396153.2
|
FAF1
|
Fas (TNFRSF6) associated factor 1
|
|
chr2_-_182545603
|
0.017
|
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1
|
|
chr12_+_65563329
|
0.016
|
ENST00000308330.2
|
LEMD3
|
LEM domain containing 3
|
|
chr9_-_123964114
|
0.015
|
ENST00000373840.4
|
RAB14
|
RAB14, member RAS oncogene family
|
|
chr11_+_119076745
|
0.013
|
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase
|
|
chr9_-_111929560
|
0.013
|
ENST00000561981.2
|
FRRS1L
|
ferric-chelate reductase 1-like
|
|
chr13_-_77900814
|
0.012
|
ENST00000544440.2
|
MYCBP2
|
MYC binding protein 2, E3 ubiquitin protein ligase
|
|
chr16_-_20911641
|
0.012
|
ENST00000324344.4
ENST00000564349.1
|
DCUN1D3
ERI2
|
DCN1, defective in cullin neddylation 1, domain containing 3
ERI1 exoribonuclease family member 2
|
|
chr9_-_37465396
|
0.011
|
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5
|
|
chr8_+_77593448
|
0.011
|
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr12_+_69864129
|
0.011
|
ENST00000547219.1
ENST00000299293.2
ENST00000549921.1
ENST00000550316.1
ENST00000548154.1
ENST00000547414.1
ENST00000550389.1
ENST00000550937.1
ENST00000549092.1
ENST00000550169.1
|
FRS2
|
fibroblast growth factor receptor substrate 2
|
|
chr15_+_79165151
|
0.010
|
ENST00000331268.5
|
MORF4L1
|
mortality factor 4 like 1
|
|
chr1_+_164528866
|
0.010
|
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr10_-_102279586
|
0.008
|
ENST00000370345.3
ENST00000451524.1
ENST00000370329.5
|
SEC31B
|
SEC31 homolog B (S. cerevisiae)
|
|
chrX_+_70586082
|
0.008
|
ENST00000373790.4
ENST00000449580.1
ENST00000423759.1
|
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa
|
|
chr21_+_34915337
|
0.008
|
ENST00000290239.6
ENST00000381692.2
ENST00000300278.4
ENST00000381679.4
|
SON
|
SON DNA binding protein
|
|
chr7_-_20256965
|
0.008
|
ENST00000400331.5
ENST00000332878.4
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr12_-_113574028
|
0.007
|
ENST00000546530.1
ENST00000261729.5
|
RASAL1
|
RAS protein activator like 1 (GAP1 like)
|
|
chr12_+_72233487
|
0.007
|
ENST00000482439.2
ENST00000550746.1
ENST00000491063.1
ENST00000319106.8
ENST00000485960.2
ENST00000393309.3
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr21_+_44394620
|
0.006
|
ENST00000291547.5
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr22_-_44258360
|
0.005
|
ENST00000330884.4
ENST00000249130.5
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr9_+_140032842
|
0.003
|
ENST00000371561.3
ENST00000315048.3
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr2_-_69870835
|
0.003
|
ENST00000409085.4
ENST00000406297.3
|
AAK1
|
AP2 associated kinase 1
|
|
chr2_-_105946491
|
0.002
|
ENST00000393359.2
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1
|
|
chr14_-_36989427
|
0.002
|
ENST00000354822.5
|
NKX2-1
|
NK2 homeobox 1
|
|
chr1_-_154155595
|
0.000
|
ENST00000328159.4
ENST00000368531.2
ENST00000323144.7
ENST00000368533.3
ENST00000341372.3
|
TPM3
|
tropomyosin 3
|
|
chr2_-_96811170
|
0.000
|
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2
|